Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Rad21_Smc3
Z-value: 2.16
Transcription factors associated with Rad21_Smc3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
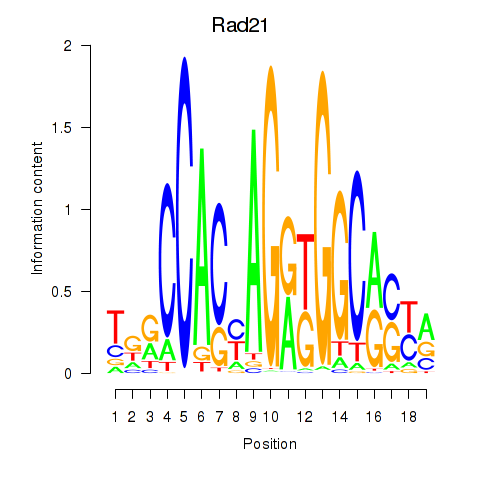
Rad21
|
ENSMUSG00000022314.11 | RAD21 cohesin complex component |
|
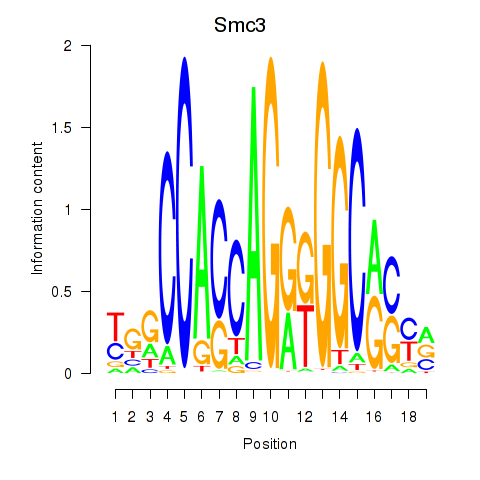
Smc3
|
ENSMUSG00000024974.11 | structural maintenance of chromosomes 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smc3 | mm39_v1_chr19_+_53588808_53588869 | -0.91 | 3.0e-02 | Click! |
| Rad21 | mm39_v1_chr15_-_51855073_51855159 | 0.90 | 4.0e-02 | Click! |
Activity profile of Rad21_Smc3 motif
Sorted Z-values of Rad21_Smc3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_126866682 | 1.68 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr6_-_52217821 | 1.60 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr16_+_16714333 | 1.55 |
ENSMUST00000027373.12
ENSMUST00000232247.2 |
Ppm1f
|
protein phosphatase 1F (PP2C domain containing) |
| chr19_-_10857734 | 1.41 |
ENSMUST00000133303.8
|
Tmem109
|
transmembrane protein 109 |
| chr8_+_22682816 | 1.36 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr1_-_133537953 | 1.26 |
ENSMUST00000164574.2
ENSMUST00000166291.8 ENSMUST00000164096.2 ENSMUST00000166915.8 |
Snrpe
|
small nuclear ribonucleoprotein E |
| chr7_-_44785603 | 1.25 |
ENSMUST00000209467.2
|
Gm45713
|
predicted gene 45713 |
| chr7_-_108769719 | 1.23 |
ENSMUST00000208136.2
ENSMUST00000036992.9 |
Lmo1
|
LIM domain only 1 |
| chr11_-_70873773 | 1.20 |
ENSMUST00000078528.7
|
C1qbp
|
complement component 1, q subcomponent binding protein |
| chr14_-_14389372 | 1.19 |
ENSMUST00000023924.4
|
Rpp14
|
ribonuclease P 14 subunit |
| chr14_+_8348779 | 1.19 |
ENSMUST00000022256.5
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr17_-_56343531 | 1.17 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr9_+_106088798 | 1.13 |
ENSMUST00000216850.2
|
Twf2
|
twinfilin actin binding protein 2 |
| chr12_-_110669076 | 1.10 |
ENSMUST00000155242.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr5_+_31078911 | 1.07 |
ENSMUST00000201571.4
|
Khk
|
ketohexokinase |
| chr6_-_85114725 | 1.06 |
ENSMUST00000174769.2
ENSMUST00000174286.3 ENSMUST00000045986.8 |
Spr
|
sepiapterin reductase |
| chr4_-_132072988 | 1.05 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr4_+_134123631 | 1.03 |
ENSMUST00000105869.9
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr8_-_108151661 | 1.01 |
ENSMUST00000003946.9
|
Nob1
|
NIN1/RPN12 binding protein 1 homolog |
| chr17_+_34423054 | 1.01 |
ENSMUST00000138491.2
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_+_24598633 | 1.00 |
ENSMUST00000138168.3
ENSMUST00000115077.8 |
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr4_-_132073048 | 0.99 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chrX_-_101232978 | 0.97 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr3_-_84063067 | 0.95 |
ENSMUST00000047368.8
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr15_-_36496880 | 0.94 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr19_+_43741550 | 0.93 |
ENSMUST00000153295.2
|
Cutc
|
cutC copper transporter |
| chr8_+_107757847 | 0.93 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr15_+_74593041 | 0.93 |
ENSMUST00000070923.3
|
Them6
|
thioesterase superfamily member 6 |
| chr5_-_117254037 | 0.92 |
ENSMUST00000086471.12
|
Suds3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr5_+_115373895 | 0.91 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr4_+_152123772 | 0.87 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr5_+_108048697 | 0.86 |
ENSMUST00000153590.2
|
Rpl5
|
ribosomal protein L5 |
| chr3_+_142266113 | 0.84 |
ENSMUST00000106221.8
|
Gbp3
|
guanylate binding protein 3 |
| chr8_-_70078152 | 0.83 |
ENSMUST00000121886.8
|
Zfp868
|
zinc finger protein 868 |
| chr4_+_127137577 | 0.81 |
ENSMUST00000142029.2
|
Smim12
|
small integral membrane protein 12 |
| chr1_+_44159106 | 0.81 |
ENSMUST00000114709.3
ENSMUST00000129068.2 |
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr2_-_66086919 | 0.81 |
ENSMUST00000125446.3
ENSMUST00000102718.10 |
Ttc21b
|
tetratricopeptide repeat domain 21B |
| chr3_-_107603778 | 0.80 |
ENSMUST00000029490.15
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1 |
| chr10_-_126866658 | 0.80 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr5_-_117254146 | 0.80 |
ENSMUST00000166397.3
|
Suds3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr16_-_10603389 | 0.79 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr17_+_25992761 | 0.77 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr3_+_96552895 | 0.75 |
ENSMUST00000119365.8
ENSMUST00000029744.6 |
Itga10
|
integrin, alpha 10 |
| chr4_-_45108038 | 0.75 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr16_+_17870724 | 0.75 |
ENSMUST00000143343.8
|
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr2_+_26476619 | 0.74 |
ENSMUST00000239075.2
ENSMUST00000174211.8 ENSMUST00000145575.10 ENSMUST00000173920.9 |
Egfl7
|
EGF-like domain 7 |
| chr4_+_124608569 | 0.73 |
ENSMUST00000030734.5
|
Sf3a3
|
splicing factor 3a, subunit 3 |
| chr7_+_34818709 | 0.73 |
ENSMUST00000205391.2
ENSMUST00000042985.11 |
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr5_-_143301113 | 0.73 |
ENSMUST00000046418.3
|
E130309D02Rik
|
RIKEN cDNA E130309D02 gene |
| chr15_+_100659622 | 0.73 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr4_+_86493905 | 0.72 |
ENSMUST00000091064.8
|
Rraga
|
Ras-related GTP binding A |
| chr11_-_3864664 | 0.72 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr4_-_126362372 | 0.71 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr5_+_124045238 | 0.70 |
ENSMUST00000023869.15
|
Denr
|
density-regulated protein |
| chr2_-_157988303 | 0.67 |
ENSMUST00000103122.10
|
Tgm2
|
transglutaminase 2, C polypeptide |
| chr13_-_55684317 | 0.67 |
ENSMUST00000021956.9
ENSMUST00000224765.2 |
Ddx41
|
DEAD box helicase 41 |
| chr1_+_161796854 | 0.66 |
ENSMUST00000160881.2
ENSMUST00000159648.2 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr6_+_124547247 | 0.66 |
ENSMUST00000184647.2
|
C1rb
|
complement component 1, r subcomponent B |
| chr7_+_16043502 | 0.65 |
ENSMUST00000002152.13
|
Bbc3
|
BCL2 binding component 3 |
| chr5_+_64317550 | 0.64 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr19_+_7394951 | 0.64 |
ENSMUST00000159348.3
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene |
| chr7_-_46360066 | 0.62 |
ENSMUST00000143082.4
|
Saal1
|
serum amyloid A-like 1 |
| chr11_-_82781369 | 0.62 |
ENSMUST00000092844.13
ENSMUST00000021033.16 ENSMUST00000018985.15 |
Rad51d
|
RAD51 paralog D |
| chr12_+_55431007 | 0.62 |
ENSMUST00000163070.8
|
Psma6
|
proteasome subunit alpha 6 |
| chr5_+_115697526 | 0.62 |
ENSMUST00000086519.12
ENSMUST00000156359.2 ENSMUST00000152976.2 |
Rplp0
|
ribosomal protein, large, P0 |
| chr4_-_141345549 | 0.61 |
ENSMUST00000053263.9
|
Tmem82
|
transmembrane protein 82 |
| chr7_-_120269462 | 0.61 |
ENSMUST00000127845.2
ENSMUST00000208635.2 ENSMUST00000033178.4 |
Pdzd9
|
PDZ domain containing 9 |
| chr12_+_24758240 | 0.60 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr3_-_5641171 | 0.60 |
ENSMUST00000071280.8
ENSMUST00000195855.6 ENSMUST00000165309.8 ENSMUST00000164828.8 |
Pex2
|
peroxisomal biogenesis factor 2 |
| chr17_-_56343625 | 0.60 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr12_+_24758724 | 0.60 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr3_+_54642957 | 0.59 |
ENSMUST00000044567.4
|
Alg5
|
asparagine-linked glycosylation 5 (dolichyl-phosphate beta-glucosyltransferase) |
| chrX_+_20529137 | 0.59 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr19_+_5012362 | 0.59 |
ENSMUST00000236917.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr16_-_20121108 | 0.58 |
ENSMUST00000048642.15
ENSMUST00000232036.2 |
Parl
|
presenilin associated, rhomboid-like |
| chr15_+_102378966 | 0.58 |
ENSMUST00000077037.13
ENSMUST00000229102.2 ENSMUST00000229618.2 ENSMUST00000229275.2 ENSMUST00000231089.2 ENSMUST00000229802.2 ENSMUST00000229854.2 ENSMUST00000108838.5 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr12_+_24758968 | 0.57 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr7_-_126625739 | 0.57 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr7_+_49428082 | 0.57 |
ENSMUST00000032715.13
|
Prmt3
|
protein arginine N-methyltransferase 3 |
| chr6_+_124489364 | 0.56 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chr5_-_114266228 | 0.56 |
ENSMUST00000053657.13
ENSMUST00000149418.2 ENSMUST00000112279.2 |
Alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr2_-_93652882 | 0.56 |
ENSMUST00000111248.8
ENSMUST00000184931.8 ENSMUST00000145838.8 ENSMUST00000028623.13 |
Ext2
|
exostosin glycosyltransferase 2 |
| chr17_+_29487881 | 0.56 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr4_+_126156118 | 0.55 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chr3_-_89008406 | 0.55 |
ENSMUST00000200659.2
|
Gm43738
|
predicted gene 43738 |
| chr12_-_26465253 | 0.55 |
ENSMUST00000020971.14
|
Rnf144a
|
ring finger protein 144A |
| chr11_+_49135018 | 0.55 |
ENSMUST00000167400.8
ENSMUST00000081794.7 |
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr19_-_8691797 | 0.55 |
ENSMUST00000206797.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr5_-_130053120 | 0.54 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr3_-_87676239 | 0.54 |
ENSMUST00000173184.2
ENSMUST00000172621.8 ENSMUST00000174759.8 ENSMUST00000172590.8 ENSMUST00000079083.12 |
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr12_-_112640626 | 0.53 |
ENSMUST00000001780.10
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr5_+_140317615 | 0.53 |
ENSMUST00000071881.10
ENSMUST00000050205.12 ENSMUST00000110827.8 |
Nudt1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr15_-_76396151 | 0.53 |
ENSMUST00000023214.11
|
Dgat1
|
diacylglycerol O-acyltransferase 1 |
| chr13_+_52000704 | 0.52 |
ENSMUST00000021903.3
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr7_-_126625657 | 0.51 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr9_-_44179348 | 0.51 |
ENSMUST00000169651.3
|
Nlrx1
|
NLR family member X1 |
| chr14_+_45567245 | 0.51 |
ENSMUST00000022380.9
|
Psmc6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr7_+_127399776 | 0.51 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr11_+_4085164 | 0.51 |
ENSMUST00000003677.11
ENSMUST00000145705.8 |
Rnf215
|
ring finger protein 215 |
| chr7_-_45084012 | 0.51 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr9_-_44179758 | 0.51 |
ENSMUST00000034621.16
ENSMUST00000168499.9 |
Nlrx1
|
NLR family member X1 |
| chr19_+_5012396 | 0.50 |
ENSMUST00000235229.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr6_+_88061464 | 0.50 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr4_+_140428777 | 0.50 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr5_+_124045552 | 0.50 |
ENSMUST00000166233.2
|
Denr
|
density-regulated protein |
| chr7_+_120442048 | 0.50 |
ENSMUST00000047875.16
|
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr4_-_43025756 | 0.50 |
ENSMUST00000098109.9
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr7_+_127399789 | 0.49 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr17_-_34109513 | 0.49 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr18_+_37898633 | 0.49 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr13_-_8921732 | 0.48 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr14_+_55842002 | 0.48 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr15_+_101184488 | 0.48 |
ENSMUST00000229525.2
ENSMUST00000230525.2 |
Atg101
|
autophagy related 101 |
| chr4_-_44072712 | 0.48 |
ENSMUST00000102936.9
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr2_+_131052283 | 0.47 |
ENSMUST00000110210.8
ENSMUST00000089506.12 ENSMUST00000110208.8 |
Ap5s1
|
adaptor-related protein 5 complex, sigma 1 subunit |
| chr4_+_138926577 | 0.47 |
ENSMUST00000145368.8
|
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr1_+_82817170 | 0.47 |
ENSMUST00000189220.7
ENSMUST00000113444.8 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr19_-_8691460 | 0.47 |
ENSMUST00000206560.2
ENSMUST00000205538.2 |
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr6_-_71376277 | 0.47 |
ENSMUST00000149415.2
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr12_+_70872266 | 0.46 |
ENSMUST00000057859.9
|
Frmd6
|
FERM domain containing 6 |
| chrX_+_158315666 | 0.46 |
ENSMUST00000057180.13
|
Bclaf3
|
Bclaf1 and Thrap3 family member 3 |
| chr19_+_6450553 | 0.46 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr6_+_59185846 | 0.45 |
ENSMUST00000062626.4
|
Tigd2
|
tigger transposable element derived 2 |
| chr6_+_108760025 | 0.44 |
ENSMUST00000032196.9
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr4_-_43025792 | 0.44 |
ENSMUST00000067481.6
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr19_+_5012336 | 0.44 |
ENSMUST00000237974.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr11_-_107080150 | 0.43 |
ENSMUST00000106757.8
ENSMUST00000018577.8 |
Nol11
|
nucleolar protein 11 |
| chr8_+_106412905 | 0.43 |
ENSMUST00000213019.2
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr12_+_113120023 | 0.42 |
ENSMUST00000049271.13
|
Tedc1
|
tubulin epsilon and delta complex 1 |
| chr10_-_10433831 | 0.42 |
ENSMUST00000019974.5
|
Rab32
|
RAB32, member RAS oncogene family |
| chr3_+_154302311 | 0.42 |
ENSMUST00000192462.6
ENSMUST00000029850.15 |
Cryz
|
crystallin, zeta |
| chr4_-_149222057 | 0.42 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr7_-_126625617 | 0.42 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr13_+_51254852 | 0.42 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr17_-_66408461 | 0.41 |
ENSMUST00000024909.15
ENSMUST00000147484.2 |
Ndufv2
|
NADH:ubiquinone oxidoreductase core subunit V2 |
| chr8_-_105261262 | 0.41 |
ENSMUST00000162466.8
ENSMUST00000034349.10 |
Nae1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr7_-_126611037 | 0.40 |
ENSMUST00000133172.2
|
Mvp
|
major vault protein |
| chr3_+_133015846 | 0.40 |
ENSMUST00000029644.16
ENSMUST00000122334.8 |
Ppa2
|
pyrophosphatase (inorganic) 2 |
| chr6_+_83078545 | 0.40 |
ENSMUST00000101254.9
|
Ccdc142
|
coiled-coil domain containing 142 |
| chr9_+_110162470 | 0.40 |
ENSMUST00000198761.5
ENSMUST00000197630.3 |
Scap
|
SREBF chaperone |
| chr8_-_41870077 | 0.40 |
ENSMUST00000033999.8
|
Frg1
|
FSHD region gene 1 |
| chr19_-_43741363 | 0.40 |
ENSMUST00000045562.6
|
Cox15
|
cytochrome c oxidase assembly protein 15 |
| chr9_+_45817795 | 0.40 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr1_+_153625243 | 0.39 |
ENSMUST00000182722.8
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr7_+_45084300 | 0.39 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr4_-_129467430 | 0.39 |
ENSMUST00000102596.8
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr11_-_69470139 | 0.39 |
ENSMUST00000048139.12
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr15_+_97682210 | 0.38 |
ENSMUST00000117892.2
ENSMUST00000229084.2 |
Slc48a1
|
solute carrier family 48 (heme transporter), member 1 |
| chr4_-_149221998 | 0.38 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr1_+_16175998 | 0.38 |
ENSMUST00000027053.8
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr11_-_62172164 | 0.38 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr3_-_116301684 | 0.38 |
ENSMUST00000153005.2
|
Rtca
|
RNA 3'-terminal phosphate cyclase |
| chr5_+_149601688 | 0.38 |
ENSMUST00000100404.6
|
B3glct
|
beta-3-glucosyltransferase |
| chr1_-_150268470 | 0.38 |
ENSMUST00000006167.13
ENSMUST00000097547.10 |
Odr4
|
odr4 GPCR localization factor homolog |
| chr8_+_70735477 | 0.38 |
ENSMUST00000087467.12
ENSMUST00000140212.8 ENSMUST00000110124.9 |
Homer3
|
homer scaffolding protein 3 |
| chrX_+_158315652 | 0.37 |
ENSMUST00000112464.8
|
Bclaf3
|
Bclaf1 and Thrap3 family member 3 |
| chr16_-_10994135 | 0.37 |
ENSMUST00000037633.16
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr9_+_71123061 | 0.37 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr1_-_92576563 | 0.37 |
ENSMUST00000053144.4
|
Otos
|
otospiralin |
| chr5_-_134205559 | 0.37 |
ENSMUST00000076228.3
|
Rcc1l
|
reculator of chromosome condensation 1 like |
| chr7_+_127400016 | 0.37 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr11_-_45835737 | 0.36 |
ENSMUST00000129820.8
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11 |
| chr3_-_105594865 | 0.36 |
ENSMUST00000090680.11
|
Ddx20
|
DEAD box helicase 20 |
| chr16_-_16950241 | 0.36 |
ENSMUST00000023453.10
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr19_-_42768374 | 0.35 |
ENSMUST00000069298.13
ENSMUST00000160455.8 ENSMUST00000162004.8 |
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr5_+_31079177 | 0.35 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chr3_-_5641295 | 0.35 |
ENSMUST00000059021.10
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr10_-_17898938 | 0.35 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chr19_+_43741513 | 0.35 |
ENSMUST00000112047.10
|
Cutc
|
cutC copper transporter |
| chr11_-_62539284 | 0.35 |
ENSMUST00000057194.9
|
Lrrc75a
|
leucine rich repeat containing 75A |
| chr4_-_44073016 | 0.34 |
ENSMUST00000128439.8
ENSMUST00000140724.3 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr11_-_100986192 | 0.34 |
ENSMUST00000019447.15
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr14_+_31887739 | 0.34 |
ENSMUST00000111994.10
ENSMUST00000168114.8 ENSMUST00000168034.8 |
Ncoa4
|
nuclear receptor coactivator 4 |
| chr16_-_84532630 | 0.34 |
ENSMUST00000116584.2
|
Mrpl39
|
mitochondrial ribosomal protein L39 |
| chr1_+_153625161 | 0.33 |
ENSMUST00000086209.10
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr10_+_24745884 | 0.33 |
ENSMUST00000092646.13
ENSMUST00000020159.15 |
Med23
|
mediator complex subunit 23 |
| chr17_+_25992742 | 0.33 |
ENSMUST00000134108.8
ENSMUST00000002350.11 |
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr9_+_107765320 | 0.33 |
ENSMUST00000191906.6
ENSMUST00000035202.4 |
Mon1a
|
MON1 homolog A, secretory traffciking associated |
| chr1_-_16590244 | 0.32 |
ENSMUST00000144138.4
ENSMUST00000145092.8 ENSMUST00000131257.9 ENSMUST00000153966.9 ENSMUST00000162435.8 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr18_+_12776358 | 0.32 |
ENSMUST00000234966.2
ENSMUST00000025294.9 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr13_-_9046020 | 0.32 |
ENSMUST00000222098.2
|
Gtpbp4
|
GTP binding protein 4 |
| chr4_+_155915729 | 0.32 |
ENSMUST00000139651.8
ENSMUST00000084097.12 |
Aurkaip1
|
aurora kinase A interacting protein 1 |
| chr2_-_152857239 | 0.31 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr3_-_120965327 | 0.31 |
ENSMUST00000170781.2
ENSMUST00000039761.12 ENSMUST00000106467.8 ENSMUST00000106466.10 ENSMUST00000164925.9 |
Rwdd3
|
RWD domain containing 3 |
| chr3_-_89905927 | 0.31 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr10_+_11157047 | 0.30 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chr1_-_38168697 | 0.30 |
ENSMUST00000027251.12
|
Rev1
|
REV1, DNA directed polymerase |
| chr5_+_120787253 | 0.30 |
ENSMUST00000156722.2
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr14_+_66534478 | 0.30 |
ENSMUST00000022623.13
|
Trim35
|
tripartite motif-containing 35 |
| chr2_+_117080212 | 0.30 |
ENSMUST00000028825.5
|
Fam98b
|
family with sequence similarity 98, member B |
| chr19_+_6450641 | 0.29 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr13_-_35211060 | 0.29 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr9_-_57552844 | 0.29 |
ENSMUST00000216979.2
ENSMUST00000034863.8 |
Csk
|
c-src tyrosine kinase |
| chr6_+_83142902 | 0.29 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr11_+_50022167 | 0.29 |
ENSMUST00000093138.13
ENSMUST00000101270.5 |
Tbc1d9b
|
TBC1 domain family, member 9B |
| chr7_+_45084257 | 0.29 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr4_+_152018142 | 0.29 |
ENSMUST00000062904.11
|
Dnajc11
|
DnaJ heat shock protein family (Hsp40) member C11 |
| chr8_-_25592001 | 0.28 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr18_-_10610048 | 0.28 |
ENSMUST00000115864.8
ENSMUST00000145320.2 ENSMUST00000097670.10 |
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr6_-_83752749 | 0.28 |
ENSMUST00000014892.8
|
Tex261
|
testis expressed gene 261 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rad21_Smc3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 1.6 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.3 | 1.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 1.0 | GO:0002481 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) cytosol to ER transport(GO:0046967) |
| 0.3 | 0.9 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.3 | 0.9 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.3 | 0.8 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.2 | 1.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 1.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 0.7 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 2.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 1.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.7 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 0.5 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.2 | 0.5 | GO:1901738 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin A metabolic process(GO:1901738) |
| 0.2 | 1.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.5 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.2 | 1.5 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 0.5 | GO:0042262 | DNA protection(GO:0042262) |
| 0.2 | 1.1 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 1.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.4 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 0.5 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 1.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 1.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 1.5 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.5 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.5 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 0.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 1.1 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 0.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.8 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.1 | 0.9 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 3.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 0.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.7 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 0.3 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 0.5 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.3 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.1 | 0.7 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 0.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.6 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 0.6 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 1.1 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.8 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.4 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.3 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 1.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 1.3 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 1.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.4 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.4 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.3 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.2 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 1.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.0 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 1.3 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 1.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.0 | 0.5 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.8 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.4 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 1.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.4 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 | 0.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.4 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 1.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.6 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.6 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.4 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.4 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.9 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.7 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.0 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 1.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 1.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 0.9 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.3 | 0.8 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 2.3 | GO:0030681 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.2 | 0.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 0.8 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 1.1 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.2 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.7 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.6 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 1.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.5 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.0 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 0.7 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.8 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0035859 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.0 | 0.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 5.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.0 | GO:0022627 | small ribosomal subunit(GO:0015935) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.5 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.4 | 1.5 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.3 | 2.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.3 | 1.0 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.3 | 0.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 1.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 1.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.2 | 0.9 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 0.7 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 1.1 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.2 | 1.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 0.6 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.2 | 0.6 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.2 | 0.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.2 | 0.5 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.2 | 0.5 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.2 | 0.5 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.2 | 1.1 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.4 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.5 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.9 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 1.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 2.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.8 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.0 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.7 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.2 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.2 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 0.5 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 3.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.3 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.0 | 0.6 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 1.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.9 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 3.6 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |