Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Rarb
Z-value: 0.28
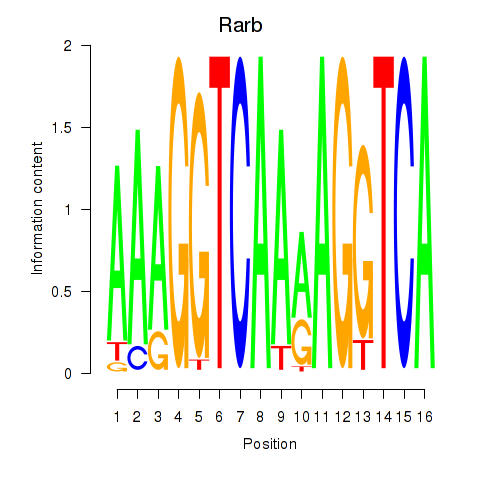
Transcription factors associated with Rarb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarb
|
ENSMUSG00000017491.10 | retinoic acid receptor, beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarb | mm39_v1_chr14_+_5894220_5894254 | -0.95 | 1.3e-02 | Click! |
Activity profile of Rarb motif
Sorted Z-values of Rarb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_126737185 | 0.14 |
ENSMUST00000168520.3
ENSMUST00000026504.13 |
Atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr9_+_56844746 | 0.10 |
ENSMUST00000034827.10
|
Imp3
|
IMP3, U3 small nucleolar ribonucleoprotein |
| chr11_-_71092282 | 0.09 |
ENSMUST00000108515.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr11_-_71092124 | 0.09 |
ENSMUST00000108514.10
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr19_-_8763771 | 0.09 |
ENSMUST00000176496.8
|
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr16_+_38167352 | 0.08 |
ENSMUST00000050273.9
ENSMUST00000120495.2 ENSMUST00000119704.2 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17, copper chaperone predicted gene 21987 |
| chr1_+_191553556 | 0.08 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr19_-_8763609 | 0.08 |
ENSMUST00000177216.8
ENSMUST00000176610.9 ENSMUST00000177056.8 |
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr2_-_163259012 | 0.07 |
ENSMUST00000127038.2
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr2_+_30171055 | 0.06 |
ENSMUST00000143119.3
|
Gm28038
|
predicted gene, 28038 |
| chr10_-_92558219 | 0.05 |
ENSMUST00000020163.7
|
Nedd1
|
neural precursor cell expressed, developmentally down-regulated gene 1 |
| chr10_+_43400074 | 0.04 |
ENSMUST00000057649.8
ENSMUST00000216543.2 |
Gm9803
|
predicted gene 9803 |
| chr1_+_4878046 | 0.03 |
ENSMUST00000027036.11
ENSMUST00000150971.8 ENSMUST00000119612.9 ENSMUST00000137887.8 ENSMUST00000115529.8 |
Lypla1
|
lysophospholipase 1 |
| chr5_+_24569802 | 0.02 |
ENSMUST00000115090.6
ENSMUST00000030834.7 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr2_+_3771709 | 0.02 |
ENSMUST00000177037.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr10_-_81332163 | 0.02 |
ENSMUST00000020463.14
|
Ncln
|
nicalin |
| chr5_-_87288177 | 0.02 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr17_+_37356872 | 0.01 |
ENSMUST00000174456.8
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr5_-_87074380 | 0.01 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr3_+_87989278 | 0.01 |
ENSMUST00000071812.11
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr11_+_105069591 | 0.01 |
ENSMUST00000106939.9
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr8_+_36956345 | 0.01 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr12_+_8027640 | 0.01 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr14_+_51648458 | 0.01 |
ENSMUST00000022438.12
|
Vmn2r88
|
vomeronasal 2, receptor 88 |
| chr4_+_156683398 | 0.01 |
ENSMUST00000074107.13
ENSMUST00000096792.8 |
Vmn2r129
|
vomeronasal 2, receptor 129 |
| chr5_-_87054796 | 0.01 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr8_+_107409684 | 0.01 |
ENSMUST00000048359.5
|
Tango6
|
transport and golgi organization 6 |
| chr13_-_25454058 | 0.01 |
ENSMUST00000057866.13
|
Nrsn1
|
neurensin 1 |
| chr11_+_83300481 | 0.01 |
ENSMUST00000175848.8
ENSMUST00000108140.10 |
Rasl10b
|
RAS-like, family 10, member B |
| chr12_+_8027767 | 0.01 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr7_+_108533613 | 0.01 |
ENSMUST00000033342.7
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr3_+_108093645 | 0.00 |
ENSMUST00000050909.7
ENSMUST00000106659.3 ENSMUST00000106656.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr2_+_154032731 | 0.00 |
ENSMUST00000081816.11
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chrX_+_53625767 | 0.00 |
ENSMUST00000169006.3
|
Gm16430
|
predicted gene 16430 |
| chr11_+_76795346 | 0.00 |
ENSMUST00000072633.4
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr14_+_33775423 | 0.00 |
ENSMUST00000058725.5
|
Antxrl
|
anthrax toxin receptor-like |
| chr12_-_75596441 | 0.00 |
ENSMUST00000218716.2
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr11_+_83299963 | 0.00 |
ENSMUST00000021022.10
|
Rasl10b
|
RAS-like, family 10, member B |
| chr7_+_35148188 | 0.00 |
ENSMUST00000118383.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr3_+_154416745 | 0.00 |
ENSMUST00000051862.8
|
Erich3
|
glutamate rich 3 |
| chr7_+_35148579 | 0.00 |
ENSMUST00000032703.10
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chrX_+_53534764 | 0.00 |
ENSMUST00000169247.3
|
Gm16405
|
predicted gene 16405 |
| chr7_+_35148461 | 0.00 |
ENSMUST00000118969.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr5_-_33375509 | 0.00 |
ENSMUST00000201475.2
|
Spon2
|
spondin 2, extracellular matrix protein |
| chr1_-_65112680 | 0.00 |
ENSMUST00000114064.3
|
Crygc
|
crystallin, gamma C |
| chr8_-_4155758 | 0.00 |
ENSMUST00000138439.2
ENSMUST00000145007.8 |
Cd209f
|
CD209f antigen |
| chr6_-_85797946 | 0.00 |
ENSMUST00000032074.5
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr1_+_24216691 | 0.00 |
ENSMUST00000054588.15
|
Col9a1
|
collagen, type IX, alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rarb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.1 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |