Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
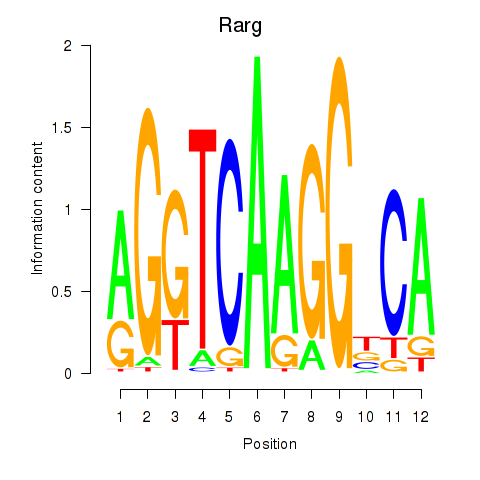
Results for Rarg
Z-value: 2.17
Transcription factors associated with Rarg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarg
|
ENSMUSG00000001288.16 | retinoic acid receptor, gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarg | mm39_v1_chr15_-_102154874_102154945 | -0.94 | 1.7e-02 | Click! |
Activity profile of Rarg motif
Sorted Z-values of Rarg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_24945423 | 1.85 |
ENSMUST00000176890.8
ENSMUST00000175689.8 |
Gmnn
|
geminin |
| chr15_+_76784110 | 0.96 |
ENSMUST00000068407.6
ENSMUST00000109793.3 |
Commd5
|
COMM domain containing 5 |
| chr16_+_32427738 | 0.96 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr16_+_32427789 | 0.90 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr2_-_126460575 | 0.86 |
ENSMUST00000028838.5
|
Hdc
|
histidine decarboxylase |
| chr17_+_24939072 | 0.85 |
ENSMUST00000054289.13
|
Rps2
|
ribosomal protein S2 |
| chr7_-_127805518 | 0.84 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr6_+_115521625 | 0.81 |
ENSMUST00000130425.8
ENSMUST00000040234.9 |
Tsen2
|
tRNA splicing endonuclease subunit 2 |
| chr2_+_179713586 | 0.81 |
ENSMUST00000108901.8
|
Mtg2
|
mitochondrial ribosome associated GTPase 2 |
| chr12_-_54250646 | 0.80 |
ENSMUST00000039516.4
|
Egln3
|
egl-9 family hypoxia-inducible factor 3 |
| chr9_-_55419442 | 0.80 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr14_-_56499690 | 0.79 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr19_+_10160249 | 0.79 |
ENSMUST00000010807.6
|
Fads1
|
fatty acid desaturase 1 |
| chr2_+_29951859 | 0.77 |
ENSMUST00000102866.10
|
Set
|
SET nuclear oncogene |
| chr19_+_10160283 | 0.77 |
ENSMUST00000235160.2
|
Fads1
|
fatty acid desaturase 1 |
| chr8_+_123789681 | 0.74 |
ENSMUST00000142541.8
ENSMUST00000125975.8 |
Spg7
|
SPG7, paraplegin matrix AAA peptidase subunit |
| chr7_-_126302315 | 0.73 |
ENSMUST00000173108.8
ENSMUST00000205515.2 |
Coro1a
|
coronin, actin binding protein 1A |
| chr15_+_101182153 | 0.73 |
ENSMUST00000048393.8
|
Atg101
|
autophagy related 101 |
| chr7_-_132178101 | 0.72 |
ENSMUST00000084500.8
|
Oat
|
ornithine aminotransferase |
| chr1_-_24044688 | 0.69 |
ENSMUST00000027338.4
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chrX_+_65692924 | 0.69 |
ENSMUST00000166241.2
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr9_+_110848339 | 0.69 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr14_-_55828511 | 0.68 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr11_+_87651359 | 0.68 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr17_+_24939225 | 0.67 |
ENSMUST00000146867.2
|
Rps2
|
ribosomal protein S2 |
| chr19_+_46695889 | 0.66 |
ENSMUST00000003655.9
|
As3mt
|
arsenite methyltransferase |
| chr8_+_72050292 | 0.65 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr11_-_96720309 | 0.65 |
ENSMUST00000167149.8
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr7_+_127345909 | 0.65 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr6_+_124690060 | 0.64 |
ENSMUST00000130279.2
|
Phb2
|
prohibitin 2 |
| chr4_-_123611974 | 0.64 |
ENSMUST00000137312.2
ENSMUST00000106206.8 |
Ndufs5
|
NADH:ubiquinone oxidoreductase core subunit S5 |
| chr2_+_24852409 | 0.64 |
ENSMUST00000028351.9
|
Dph7
|
diphthamine biosynethesis 7 |
| chr5_-_110927803 | 0.63 |
ENSMUST00000112426.8
|
Pus1
|
pseudouridine synthase 1 |
| chr5_-_45607485 | 0.62 |
ENSMUST00000154962.8
ENSMUST00000118097.8 ENSMUST00000198258.5 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr11_-_117673008 | 0.62 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr17_-_29483075 | 0.62 |
ENSMUST00000024802.10
|
Ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr18_-_36903237 | 0.62 |
ENSMUST00000235494.2
|
Hars
|
histidyl-tRNA synthetase |
| chr4_-_116508842 | 0.60 |
ENSMUST00000030455.15
|
Akr1a1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr12_+_76353835 | 0.60 |
ENSMUST00000220321.2
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr17_-_74630882 | 0.59 |
ENSMUST00000164832.9
|
Dpy30
|
dpy-30, histone methyltransferase complex regulatory subunit |
| chr5_-_122959321 | 0.59 |
ENSMUST00000197074.5
ENSMUST00000199406.5 ENSMUST00000196640.5 ENSMUST00000197719.5 ENSMUST00000200645.5 |
Anapc5
|
anaphase-promoting complex subunit 5 |
| chr7_-_133384449 | 0.59 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr9_+_73009680 | 0.58 |
ENSMUST00000034737.13
ENSMUST00000173734.9 ENSMUST00000167514.2 ENSMUST00000174203.3 |
Khdc3
Gm20509
|
KH domain containing 3, subcortical maternal complex member predicted gene 20509 |
| chr3_-_89008406 | 0.58 |
ENSMUST00000200659.2
|
Gm43738
|
predicted gene 43738 |
| chr13_-_30168374 | 0.58 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
| chr18_-_43610829 | 0.57 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chrX_-_100312629 | 0.57 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr2_+_32477069 | 0.57 |
ENSMUST00000102818.11
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr6_-_52217821 | 0.57 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr7_-_24672055 | 0.57 |
ENSMUST00000205871.2
|
Rabac1
|
Rab acceptor 1 (prenylated) |
| chr4_-_87724533 | 0.56 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr15_+_6451721 | 0.56 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr7_-_144837719 | 0.56 |
ENSMUST00000058022.6
|
Tpcn2
|
two pore segment channel 2 |
| chr9_-_106324642 | 0.56 |
ENSMUST00000185334.7
ENSMUST00000187001.2 ENSMUST00000171678.9 ENSMUST00000190798.7 ENSMUST00000048685.13 ENSMUST00000171925.8 |
Abhd14a
|
abhydrolase domain containing 14A |
| chr11_-_84807164 | 0.55 |
ENSMUST00000103195.5
|
Znhit3
|
zinc finger, HIT type 3 |
| chr6_+_121187636 | 0.55 |
ENSMUST00000032233.9
|
Tuba8
|
tubulin, alpha 8 |
| chr12_-_71183371 | 0.55 |
ENSMUST00000221367.2
ENSMUST00000220482.2 ENSMUST00000221892.2 ENSMUST00000221178.2 ENSMUST00000221559.2 ENSMUST00000166120.9 ENSMUST00000021486.10 ENSMUST00000221797.2 ENSMUST00000221815.2 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr5_-_45607463 | 0.55 |
ENSMUST00000197946.5
ENSMUST00000127562.3 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr7_+_43321426 | 0.55 |
ENSMUST00000038332.9
|
Ctu1
|
cytosolic thiouridylase subunit 1 |
| chr17_-_34219225 | 0.55 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr14_+_43951187 | 0.54 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr2_+_180098026 | 0.54 |
ENSMUST00000038259.13
|
Slco4a1
|
solute carrier organic anion transporter family, member 4a1 |
| chr17_+_29333116 | 0.54 |
ENSMUST00000233717.2
ENSMUST00000141239.2 |
Rab44
|
RAB44, member RAS oncogene family |
| chr8_+_71261073 | 0.54 |
ENSMUST00000000808.8
ENSMUST00000212657.2 ENSMUST00000212146.2 |
Il12rb1
|
interleukin 12 receptor, beta 1 |
| chr9_+_65370077 | 0.54 |
ENSMUST00000215170.2
|
Spg21
|
SPG21, maspardin |
| chr1_+_63769772 | 0.53 |
ENSMUST00000027103.7
|
Fastkd2
|
FAST kinase domains 2 |
| chr6_-_85114725 | 0.53 |
ENSMUST00000174769.2
ENSMUST00000174286.3 ENSMUST00000045986.8 |
Spr
|
sepiapterin reductase |
| chr5_+_122344854 | 0.53 |
ENSMUST00000145854.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr17_+_48539782 | 0.53 |
ENSMUST00000113251.10
ENSMUST00000048782.7 |
Trem1
|
triggering receptor expressed on myeloid cells 1 |
| chr9_-_118986123 | 0.52 |
ENSMUST00000010795.5
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr10_-_62178453 | 0.52 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr3_-_83947416 | 0.52 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chr1_-_63215812 | 0.52 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr12_-_115122455 | 0.52 |
ENSMUST00000103523.2
|
Ighv1-53
|
immunoglobulin heavy variable 1-53 |
| chr16_-_22946441 | 0.51 |
ENSMUST00000133847.9
ENSMUST00000115338.8 ENSMUST00000023598.15 |
Rfc4
|
replication factor C (activator 1) 4 |
| chr10_+_80008768 | 0.50 |
ENSMUST00000041882.7
|
Fam174c
|
family with sequence similarity 174, member C |
| chr10_+_61010983 | 0.50 |
ENSMUST00000143207.8
ENSMUST00000148181.8 ENSMUST00000151886.8 |
Tbata
|
thymus, brain and testes associated |
| chr13_+_73476629 | 0.50 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr3_+_159545309 | 0.50 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr3_-_116474909 | 0.50 |
ENSMUST00000140672.3
|
Gm43191
|
predicted gene 43191 |
| chr6_-_124410452 | 0.50 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr7_-_28441173 | 0.50 |
ENSMUST00000171183.2
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr8_+_70980374 | 0.50 |
ENSMUST00000119353.9
ENSMUST00000075491.14 ENSMUST00000119698.8 |
Fkbp8
|
FK506 binding protein 8 |
| chr11_+_32233511 | 0.50 |
ENSMUST00000093209.4
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chrX_+_52076998 | 0.50 |
ENSMUST00000026723.9
|
Hprt
|
hypoxanthine guanine phosphoribosyl transferase |
| chr19_+_34194990 | 0.49 |
ENSMUST00000119603.2
|
Stambpl1
|
STAM binding protein like 1 |
| chr7_-_121700958 | 0.48 |
ENSMUST00000139456.2
ENSMUST00000106471.9 ENSMUST00000123296.8 ENSMUST00000033157.10 |
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr11_+_51152546 | 0.48 |
ENSMUST00000130641.8
|
Clk4
|
CDC like kinase 4 |
| chr8_+_75836187 | 0.48 |
ENSMUST00000164309.3
ENSMUST00000212426.2 ENSMUST00000212811.2 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr17_-_26016039 | 0.47 |
ENSMUST00000165838.9
ENSMUST00000002344.7 |
Metrn
|
meteorin, glial cell differentiation regulator |
| chr2_-_174305856 | 0.47 |
ENSMUST00000016396.8
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr8_+_75742850 | 0.47 |
ENSMUST00000109940.2
|
Hmgxb4
|
HMG box domain containing 4 |
| chr14_-_73613385 | 0.47 |
ENSMUST00000227454.2
|
Itm2b
|
integral membrane protein 2B |
| chr8_-_11685726 | 0.47 |
ENSMUST00000033905.13
ENSMUST00000169782.3 |
Ankrd10
|
ankyrin repeat domain 10 |
| chr2_+_92205651 | 0.46 |
ENSMUST00000028650.9
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr9_+_20914211 | 0.46 |
ENSMUST00000214124.2
ENSMUST00000216818.2 |
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr2_-_32594156 | 0.46 |
ENSMUST00000127812.3
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr19_+_42158995 | 0.46 |
ENSMUST00000169536.8
ENSMUST00000099443.11 |
Zfyve27
|
zinc finger, FYVE domain containing 27 |
| chr17_-_45903410 | 0.46 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr11_+_96941637 | 0.45 |
ENSMUST00000168565.2
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr11_-_60702081 | 0.45 |
ENSMUST00000018744.15
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr5_-_110928436 | 0.45 |
ENSMUST00000149208.2
ENSMUST00000031483.15 ENSMUST00000086643.12 ENSMUST00000170468.8 ENSMUST00000031481.13 |
Pus1
|
pseudouridine synthase 1 |
| chr2_+_180222985 | 0.44 |
ENSMUST00000169630.8
|
Mrgbp
|
MRG/MORF4L binding protein |
| chr5_+_137756407 | 0.44 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr10_+_77418221 | 0.44 |
ENSMUST00000162943.9
|
Pttg1ip
|
pituitary tumor-transforming 1 interacting protein |
| chr5_-_139330906 | 0.44 |
ENSMUST00000049630.13
|
Cox19
|
cytochrome c oxidase assembly protein 19 |
| chr17_+_25992761 | 0.44 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr5_-_122959361 | 0.44 |
ENSMUST00000086216.9
|
Anapc5
|
anaphase-promoting complex subunit 5 |
| chr17_-_44416665 | 0.44 |
ENSMUST00000024757.14
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr11_+_100513747 | 0.44 |
ENSMUST00000142993.2
|
Nkiras2
|
NFKB inhibitor interacting Ras-like protein 2 |
| chr12_+_83997382 | 0.43 |
ENSMUST00000053744.9
|
Riox1
|
ribosomal oxygenase 1 |
| chr7_+_28136861 | 0.42 |
ENSMUST00000108292.9
ENSMUST00000108289.8 |
Gmfg
|
glia maturation factor, gamma |
| chr4_-_87724512 | 0.42 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr8_-_73324877 | 0.42 |
ENSMUST00000058733.9
ENSMUST00000167290.8 |
Smim7
|
small integral membrane protein 7 |
| chr10_-_71180763 | 0.42 |
ENSMUST00000045887.9
|
Cisd1
|
CDGSH iron sulfur domain 1 |
| chr11_+_80320558 | 0.42 |
ENSMUST00000173565.2
|
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr15_+_76227695 | 0.42 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr2_+_103900164 | 0.42 |
ENSMUST00000111131.9
ENSMUST00000111132.8 ENSMUST00000129749.8 |
Cd59b
|
CD59b antigen |
| chr5_+_73648368 | 0.41 |
ENSMUST00000113558.8
ENSMUST00000063882.12 |
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr7_-_30855295 | 0.41 |
ENSMUST00000186723.3
|
Gramd1a
|
GRAM domain containing 1A |
| chr9_+_108765701 | 0.41 |
ENSMUST00000026743.14
ENSMUST00000194047.3 |
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr4_-_126094910 | 0.41 |
ENSMUST00000136157.8
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr1_+_40123858 | 0.41 |
ENSMUST00000027243.13
|
Il1r2
|
interleukin 1 receptor, type II |
| chr4_-_107928567 | 0.41 |
ENSMUST00000106701.2
|
Scp2
|
sterol carrier protein 2, liver |
| chr14_+_76725876 | 0.41 |
ENSMUST00000101618.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr4_+_108336164 | 0.41 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr9_+_21634779 | 0.41 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr10_+_128073900 | 0.40 |
ENSMUST00000105245.3
|
Timeless
|
timeless circadian clock 1 |
| chr6_-_146536025 | 0.40 |
ENSMUST00000037709.16
|
Tm7sf3
|
transmembrane 7 superfamily member 3 |
| chr18_-_35795233 | 0.40 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr7_+_141056305 | 0.40 |
ENSMUST00000117634.2
|
Tspan4
|
tetraspanin 4 |
| chr17_+_26471870 | 0.40 |
ENSMUST00000025023.15
|
Luc7l
|
Luc7-like |
| chr6_-_120508250 | 0.40 |
ENSMUST00000075303.7
|
Hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr9_+_110248815 | 0.39 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr1_+_135693818 | 0.39 |
ENSMUST00000038945.6
|
Phlda3
|
pleckstrin homology like domain, family A, member 3 |
| chr7_+_126446588 | 0.39 |
ENSMUST00000141805.8
ENSMUST00000064110.14 ENSMUST00000205938.2 ENSMUST00000152051.8 |
Doc2a
|
double C2, alpha |
| chr2_+_118692435 | 0.39 |
ENSMUST00000028807.6
|
Ivd
|
isovaleryl coenzyme A dehydrogenase |
| chr5_-_137074880 | 0.39 |
ENSMUST00000144303.2
ENSMUST00000111080.8 |
Ap1s1
|
adaptor protein complex AP-1, sigma 1 |
| chr14_+_66043281 | 0.38 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr4_-_116664729 | 0.38 |
ENSMUST00000106455.8
ENSMUST00000030451.10 |
Toe1
|
target of EGR1, member 1 (nuclear) |
| chr9_+_21634918 | 0.38 |
ENSMUST00000213114.2
|
Ldlr
|
low density lipoprotein receptor |
| chr6_+_58617521 | 0.38 |
ENSMUST00000145161.8
ENSMUST00000203146.3 ENSMUST00000114294.8 ENSMUST00000204948.2 |
Abcg2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr15_+_34238174 | 0.38 |
ENSMUST00000022867.5
ENSMUST00000226627.2 |
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr17_+_24851647 | 0.38 |
ENSMUST00000047611.4
|
Nthl1
|
nth (endonuclease III)-like 1 (E.coli) |
| chr5_-_30278552 | 0.38 |
ENSMUST00000198095.2
ENSMUST00000196872.2 ENSMUST00000026846.11 |
Tyms
|
thymidylate synthase |
| chr16_-_55858089 | 0.37 |
ENSMUST00000059052.9
|
Trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr7_+_15853792 | 0.37 |
ENSMUST00000006178.5
|
Kptn
|
kaptin |
| chr8_+_85696695 | 0.37 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr11_-_51579441 | 0.37 |
ENSMUST00000007921.9
|
0610009B22Rik
|
RIKEN cDNA 0610009B22 gene |
| chr7_-_141934524 | 0.37 |
ENSMUST00000209263.2
|
Gm49369
|
predicted gene, 49369 |
| chr17_+_26471889 | 0.37 |
ENSMUST00000114976.9
ENSMUST00000140427.8 ENSMUST00000119928.8 |
Luc7l
|
Luc7-like |
| chr11_+_51654511 | 0.37 |
ENSMUST00000020653.6
|
Sar1b
|
secretion associated Ras related GTPase 1B |
| chr6_+_113600920 | 0.37 |
ENSMUST00000035673.8
|
Vhl
|
von Hippel-Lindau tumor suppressor |
| chr13_-_56283331 | 0.37 |
ENSMUST00000045788.9
ENSMUST00000016081.13 |
Macroh2a1
|
macroH2A.1 histone |
| chr7_-_24672083 | 0.37 |
ENSMUST00000076961.9
|
Rabac1
|
Rab acceptor 1 (prenylated) |
| chr9_+_98178608 | 0.37 |
ENSMUST00000112935.8
|
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr2_-_155668567 | 0.37 |
ENSMUST00000109638.2
ENSMUST00000134278.2 |
Eif6
|
eukaryotic translation initiation factor 6 |
| chr3_-_105867415 | 0.37 |
ENSMUST00000118209.8
|
Atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr11_-_96720738 | 0.37 |
ENSMUST00000107657.8
ENSMUST00000081775.12 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr13_-_74465353 | 0.36 |
ENSMUST00000022060.7
|
Pdcd6
|
programmed cell death 6 |
| chr6_+_58810674 | 0.36 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr9_+_62754252 | 0.36 |
ENSMUST00000124984.2
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr17_-_35984409 | 0.36 |
ENSMUST00000162266.8
ENSMUST00000160734.8 ENSMUST00000159852.2 ENSMUST00000160039.8 |
Gtf2h4
|
general transcription factor II H, polypeptide 4 |
| chr4_-_129436465 | 0.36 |
ENSMUST00000102597.5
|
Hdac1
|
histone deacetylase 1 |
| chr16_+_17328924 | 0.36 |
ENSMUST00000232372.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr3_+_146110387 | 0.36 |
ENSMUST00000106151.8
ENSMUST00000106153.9 ENSMUST00000039021.11 ENSMUST00000106149.8 ENSMUST00000149262.8 |
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr16_+_32002750 | 0.36 |
ENSMUST00000231512.2
|
Bex6
|
brain expressed family member 6 |
| chr17_-_26011357 | 0.36 |
ENSMUST00000236683.2
|
Antkmt
|
adenine nucleotide translocase lysine methyltransferase |
| chr11_-_113456568 | 0.36 |
ENSMUST00000071539.10
ENSMUST00000106633.10 ENSMUST00000042657.16 ENSMUST00000149034.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr6_+_35154319 | 0.35 |
ENSMUST00000201374.4
ENSMUST00000043815.16 |
Nup205
|
nucleoporin 205 |
| chr9_+_98178646 | 0.35 |
ENSMUST00000112938.8
ENSMUST00000112937.3 |
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr9_-_96360676 | 0.35 |
ENSMUST00000057500.6
|
Rnf7
|
ring finger protein 7 |
| chr14_-_55873026 | 0.35 |
ENSMUST00000125133.2
ENSMUST00000047131.16 |
Ipo4
|
importin 4 |
| chr16_+_33614378 | 0.35 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr17_-_34962823 | 0.35 |
ENSMUST00000069507.9
|
C4b
|
complement component 4B (Chido blood group) |
| chr10_+_79650496 | 0.35 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr19_-_60862964 | 0.34 |
ENSMUST00000025961.7
|
Prdx3
|
peroxiredoxin 3 |
| chr9_-_57375269 | 0.34 |
ENSMUST00000215059.2
ENSMUST00000046587.8 ENSMUST00000214256.2 |
Scamp5
|
secretory carrier membrane protein 5 |
| chr8_-_127672590 | 0.34 |
ENSMUST00000179857.3
ENSMUST00000212771.2 |
Tomm20
|
translocase of outer mitochondrial membrane 20 |
| chr17_+_29900144 | 0.34 |
ENSMUST00000127695.2
|
Cmtr1
|
cap methyltransferase 1 |
| chr9_+_20927271 | 0.34 |
ENSMUST00000086399.6
|
Icam1
|
intercellular adhesion molecule 1 |
| chr4_-_116485118 | 0.34 |
ENSMUST00000030456.14
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr2_-_25136857 | 0.34 |
ENSMUST00000228627.3
|
Ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr7_+_51537645 | 0.34 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr1_+_181952302 | 0.34 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr5_-_115622356 | 0.33 |
ENSMUST00000112067.8
|
Sirt4
|
sirtuin 4 |
| chrX_+_36059274 | 0.33 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr12_+_83572774 | 0.33 |
ENSMUST00000223291.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr6_-_48747767 | 0.33 |
ENSMUST00000204036.2
|
Gimap3
|
GTPase, IMAP family member 3 |
| chr10_+_78410803 | 0.33 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr10_+_82534841 | 0.33 |
ENSMUST00000020478.14
|
Hcfc2
|
host cell factor C2 |
| chr9_+_4376556 | 0.33 |
ENSMUST00000212075.2
|
Msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr9_-_20871081 | 0.32 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr2_+_29951766 | 0.32 |
ENSMUST00000149578.8
|
Set
|
SET nuclear oncogene |
| chr15_+_10981833 | 0.32 |
ENSMUST00000070877.7
|
Amacr
|
alpha-methylacyl-CoA racemase |
| chr2_+_19376447 | 0.32 |
ENSMUST00000023856.9
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr5_-_34794185 | 0.32 |
ENSMUST00000149657.5
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr1_+_172139934 | 0.31 |
ENSMUST00000039506.15
|
Igsf8
|
immunoglobulin superfamily, member 8 |
| chrX_-_35909011 | 0.31 |
ENSMUST00000051906.13
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr10_+_80905869 | 0.31 |
ENSMUST00000005057.7
|
Thop1
|
thimet oligopeptidase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rarg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.2 | 0.8 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 0.8 | GO:1905167 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 0.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 0.9 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.2 | 0.6 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.7 | GO:0042710 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.2 | 0.9 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.2 | 1.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.2 | 0.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.5 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.2 | 0.5 | GO:0046101 | GMP catabolic process(GO:0046038) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 | 0.8 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 0.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.6 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.1 | 0.4 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.4 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 0.4 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.1 | 0.5 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 0.1 | 0.9 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.1 | 0.4 | GO:0006296 | base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 0.4 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.4 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.7 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.3 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.5 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 0.5 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 1.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.4 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.4 | GO:0032385 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.4 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 1.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.6 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.4 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.1 | 0.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.5 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.1 | 0.7 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.7 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.3 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 1.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.4 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.4 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.7 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.6 | GO:0060744 | positive regulation of exit from mitosis(GO:0031536) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.9 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.2 | GO:0046203 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) spermidine catabolic process(GO:0046203) |
| 0.1 | 0.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.5 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.5 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 0.3 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.3 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 1.0 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.2 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.1 | 0.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.5 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0061198 | fungiform papilla formation(GO:0061198) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.4 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.4 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.5 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.3 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.3 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.1 | 0.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.2 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 1.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.7 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.5 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.3 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.3 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.3 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.3 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.3 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.3 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.1 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.2 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.4 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 0.6 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.0 | 0.1 | GO:0035854 | basophil differentiation(GO:0030221) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.2 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.5 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0070627 | ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.5 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.4 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.0 | 0.3 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.0 | GO:1904173 | regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.0 | 0.1 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0019243 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.0 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.5 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 1.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.3 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.6 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.6 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.5 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.3 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0045019 | negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.4 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.3 | 1.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.2 | 0.8 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 0.6 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 0.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 2.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.6 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 0.7 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.5 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.4 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.1 | 0.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.3 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.6 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 2.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 1.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 1.8 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.5 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.4 | 1.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.4 | 1.9 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 0.6 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.2 | 0.6 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.2 | 0.7 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.2 | 0.5 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.2 | 0.7 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 0.5 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.2 | 0.5 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.4 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.4 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 0.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.7 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.4 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.3 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.4 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.8 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.4 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 1.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.3 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.3 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.8 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.1 | 0.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.5 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.2 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.1 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.3 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.4 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 1.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.0 | 1.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 1.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 2.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 3.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 4.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 3.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |