Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
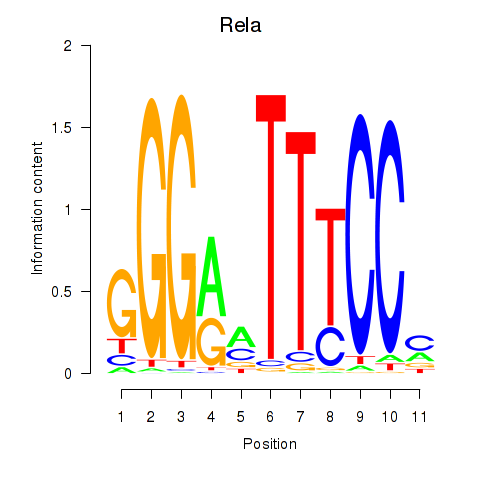
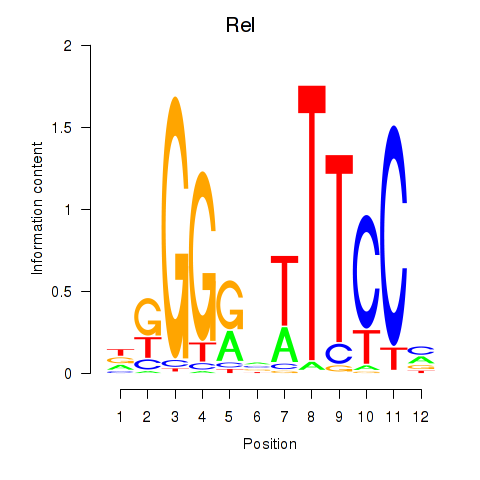
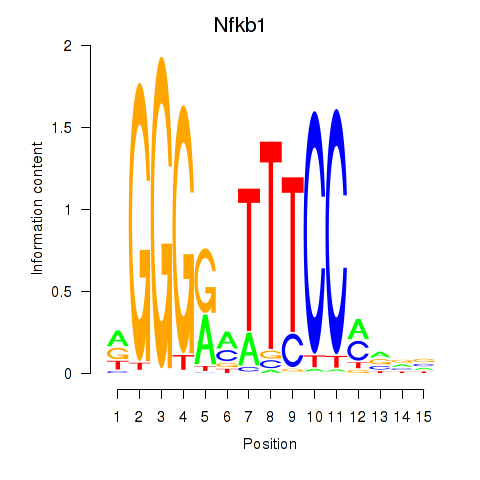
Results for Rela_Rel_Nfkb1
Z-value: 0.42
Transcription factors associated with Rela_Rel_Nfkb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rela
|
ENSMUSG00000024927.9 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) |
|
Rel
|
ENSMUSG00000020275.10 | reticuloendotheliosis oncogene |
|
Nfkb1
|
ENSMUSG00000028163.18 | nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb1 | mm39_v1_chr3_-_135373560_135373582 | 0.86 | 6.4e-02 | Click! |
| Rel | mm39_v1_chr11_-_23720953_23721028 | 0.49 | 4.0e-01 | Click! |
| Rela | mm39_v1_chr19_+_5687503_5687514 | -0.03 | 9.6e-01 | Click! |
Activity profile of Rela_Rel_Nfkb1 motif
Sorted Z-values of Rela_Rel_Nfkb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_35658131 | 0.36 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr6_-_129252396 | 0.29 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr12_-_55539372 | 0.29 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr6_-_129252323 | 0.23 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr7_-_25488060 | 0.16 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr9_-_7872970 | 0.16 |
ENSMUST00000115672.2
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr18_-_4352944 | 0.16 |
ENSMUST00000025078.10
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr16_-_18052937 | 0.16 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr8_+_72128920 | 0.15 |
ENSMUST00000110013.10
ENSMUST00000051995.14 |
Jak3
|
Janus kinase 3 |
| chr12_-_80307110 | 0.15 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr7_-_19363280 | 0.15 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr3_+_142202642 | 0.14 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr11_-_69771797 | 0.13 |
ENSMUST00000238978.2
|
Kctd11
|
potassium channel tetramerisation domain containing 11 |
| chr17_-_23964807 | 0.13 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr5_+_90907207 | 0.12 |
ENSMUST00000031318.6
|
Cxcl5
|
chemokine (C-X-C motif) ligand 5 |
| chr5_-_92496730 | 0.11 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr4_+_136013372 | 0.11 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr4_+_63280675 | 0.11 |
ENSMUST00000075341.4
|
Orm2
|
orosomucoid 2 |
| chr11_+_70431063 | 0.10 |
ENSMUST00000018429.12
ENSMUST00000108557.10 ENSMUST00000108556.2 |
Pld2
|
phospholipase D2 |
| chr7_-_126620378 | 0.10 |
ENSMUST00000159916.5
|
Prrt2
|
proline-rich transmembrane protein 2 |
| chr10_-_18891095 | 0.10 |
ENSMUST00000019997.11
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr17_+_35598583 | 0.09 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr7_-_103320398 | 0.09 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr17_+_35643818 | 0.08 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr7_+_30121147 | 0.08 |
ENSMUST00000108176.9
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr12_-_84240781 | 0.08 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr9_+_32607301 | 0.08 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr8_-_25592385 | 0.08 |
ENSMUST00000064883.14
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr12_-_40298072 | 0.07 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr4_-_40239778 | 0.07 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr11_-_109188917 | 0.07 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr12_+_111132847 | 0.07 |
ENSMUST00000021706.11
|
Traf3
|
TNF receptor-associated factor 3 |
| chr7_+_99184645 | 0.07 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr17_+_34258411 | 0.07 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr5_-_105287405 | 0.07 |
ENSMUST00000100961.5
ENSMUST00000031235.13 ENSMUST00000197799.2 ENSMUST00000199629.2 ENSMUST00000196677.5 ENSMUST00000100962.8 |
Gbp9
Gbp8
Gbp4
|
guanylate-binding protein 9 guanylate-binding protein 8 guanylate binding protein 4 |
| chr3_+_86131970 | 0.07 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr17_-_24424456 | 0.07 |
ENSMUST00000201583.2
ENSMUST00000202925.4 ENSMUST00000167791.9 ENSMUST00000201960.4 ENSMUST00000040474.11 ENSMUST00000201089.4 ENSMUST00000201301.4 ENSMUST00000201805.4 ENSMUST00000168410.9 ENSMUST00000097376.10 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr1_-_65142521 | 0.07 |
ENSMUST00000061497.9
|
Cryga
|
crystallin, gamma A |
| chr17_+_35643853 | 0.07 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr5_-_132570710 | 0.06 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr2_-_37312881 | 0.06 |
ENSMUST00000112936.4
ENSMUST00000112934.8 |
Rc3h2
|
ring finger and CCCH-type zinc finger domains 2 |
| chr8_-_105357941 | 0.06 |
ENSMUST00000034351.8
|
Rrad
|
Ras-related associated with diabetes |
| chr2_-_26184450 | 0.06 |
ENSMUST00000217256.2
ENSMUST00000227200.2 |
Ccdc187
|
coiled-coil domain containing 187 |
| chr11_-_23720953 | 0.06 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr10_-_79940168 | 0.06 |
ENSMUST00000219260.2
|
Sbno2
|
strawberry notch 2 |
| chr5_-_92511136 | 0.06 |
ENSMUST00000077820.6
|
Cxcl11
|
chemokine (C-X-C motif) ligand 11 |
| chr8_-_25592001 | 0.06 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr3_-_126792056 | 0.06 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr7_+_99184858 | 0.06 |
ENSMUST00000032995.15
ENSMUST00000162404.8 |
Arrb1
|
arrestin, beta 1 |
| chr11_+_101066867 | 0.06 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr1_-_36312482 | 0.06 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr4_+_63262775 | 0.06 |
ENSMUST00000030044.3
|
Orm1
|
orosomucoid 1 |
| chr2_+_62494622 | 0.06 |
ENSMUST00000028257.3
|
Gca
|
grancalcin |
| chr9_-_14526246 | 0.06 |
ENSMUST00000013220.8
|
Amotl1
|
angiomotin-like 1 |
| chr12_-_35584968 | 0.05 |
ENSMUST00000116436.9
|
Ahr
|
aryl-hydrocarbon receptor |
| chr15_+_98065039 | 0.05 |
ENSMUST00000031914.6
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr18_+_4994600 | 0.05 |
ENSMUST00000140448.8
|
Svil
|
supervillin |
| chr7_-_44465043 | 0.05 |
ENSMUST00000107893.9
|
Atf5
|
activating transcription factor 5 |
| chr17_-_56312555 | 0.05 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr3_+_14706781 | 0.05 |
ENSMUST00000029071.9
|
Car13
|
carbonic anhydrase 13 |
| chr19_+_3372296 | 0.05 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr12_+_111132779 | 0.05 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3 |
| chr15_-_38300937 | 0.05 |
ENSMUST00000227920.2
ENSMUST00000074043.7 ENSMUST00000228416.2 |
Klf10
|
Kruppel-like factor 10 |
| chr7_+_30121776 | 0.05 |
ENSMUST00000108175.8
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr19_-_42740898 | 0.05 |
ENSMUST00000237747.2
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr17_-_84495364 | 0.05 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr13_+_42205491 | 0.05 |
ENSMUST00000060148.6
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr1_+_74193138 | 0.05 |
ENSMUST00000027372.8
ENSMUST00000106899.4 |
Cxcr2
|
chemokine (C-X-C motif) receptor 2 |
| chr12_-_80306865 | 0.05 |
ENSMUST00000167327.2
|
Actn1
|
actinin, alpha 1 |
| chr4_-_40239700 | 0.05 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr8_+_72050292 | 0.05 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr12_+_79177523 | 0.05 |
ENSMUST00000021550.7
|
Arg2
|
arginase type II |
| chr5_-_92475927 | 0.04 |
ENSMUST00000113093.5
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr19_-_11852453 | 0.04 |
ENSMUST00000213954.2
ENSMUST00000217617.2 |
Olfr1419
|
olfactory receptor 1419 |
| chr5_-_105441554 | 0.04 |
ENSMUST00000050011.10
ENSMUST00000196520.2 |
Gm43302
Gbp6
|
predicted gene 43302 guanylate binding protein 6 |
| chr7_-_127048280 | 0.04 |
ENSMUST00000053392.11
|
Zfp689
|
zinc finger protein 689 |
| chr2_-_152672535 | 0.04 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr16_-_55659194 | 0.04 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr7_+_29467971 | 0.04 |
ENSMUST00000032802.5
|
Zfp84
|
zinc finger protein 84 |
| chr8_-_25591737 | 0.04 |
ENSMUST00000098866.11
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr5_-_121975676 | 0.04 |
ENSMUST00000197892.3
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr19_-_10582773 | 0.04 |
ENSMUST00000237788.2
|
Tkfc
|
triokinase, FMN cyclase |
| chr2_-_86926352 | 0.04 |
ENSMUST00000217066.3
ENSMUST00000214636.2 |
Olfr1109
|
olfactory receptor 1109 |
| chr14_+_20344765 | 0.04 |
ENSMUST00000223663.2
ENSMUST00000022343.6 ENSMUST00000224066.2 ENSMUST00000223941.2 ENSMUST00000224311.2 |
Nudt13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr4_+_112089442 | 0.04 |
ENSMUST00000038455.12
ENSMUST00000170945.2 |
Skint3
|
selection and upkeep of intraepithelial T cells 3 |
| chr9_+_64718596 | 0.04 |
ENSMUST00000038890.6
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr9_+_113570580 | 0.04 |
ENSMUST00000216817.2
|
Clasp2
|
CLIP associating protein 2 |
| chr1_+_36346824 | 0.04 |
ENSMUST00000142319.8
ENSMUST00000097778.9 ENSMUST00000115031.8 ENSMUST00000115032.8 ENSMUST00000137906.2 ENSMUST00000115029.2 |
Arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr7_-_102774821 | 0.04 |
ENSMUST00000211075.3
ENSMUST00000215304.2 ENSMUST00000213281.2 |
Olfr586
|
olfactory receptor 586 |
| chr11_-_34048538 | 0.04 |
ENSMUST00000223852.2
|
4930469K13Rik
|
RIKEN cDNA 4930469K13 gene |
| chr15_+_6599001 | 0.04 |
ENSMUST00000227175.2
|
Fyb
|
FYN binding protein |
| chr9_+_110709223 | 0.04 |
ENSMUST00000084926.9
|
Als2cl
|
ALS2 C-terminal like |
| chr14_+_56122404 | 0.04 |
ENSMUST00000022831.5
|
Khnyn
|
KH and NYN domain containing |
| chr9_-_7873017 | 0.04 |
ENSMUST00000013949.15
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr1_+_186947934 | 0.04 |
ENSMUST00000160471.8
|
Gpatch2
|
G patch domain containing 2 |
| chr9_-_72399221 | 0.04 |
ENSMUST00000185151.8
ENSMUST00000085358.12 ENSMUST00000184125.8 ENSMUST00000183574.8 ENSMUST00000184831.8 |
Tex9
|
testis expressed gene 9 |
| chr7_+_100757975 | 0.04 |
ENSMUST00000098250.10
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr11_+_77576981 | 0.04 |
ENSMUST00000100802.11
ENSMUST00000181023.2 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr16_+_32734464 | 0.04 |
ENSMUST00000023491.13
ENSMUST00000170899.8 ENSMUST00000170201.8 ENSMUST00000165616.8 ENSMUST00000135193.9 |
Lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr18_+_69726654 | 0.04 |
ENSMUST00000200921.4
|
Tcf4
|
transcription factor 4 |
| chr7_-_126483851 | 0.04 |
ENSMUST00000071268.11
ENSMUST00000117394.2 |
Taok2
|
TAO kinase 2 |
| chr19_+_46294119 | 0.04 |
ENSMUST00000111881.4
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr16_-_32876544 | 0.04 |
ENSMUST00000115100.9
|
Iqcg
|
IQ motif containing G |
| chr2_-_38816229 | 0.04 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr8_-_48443525 | 0.04 |
ENSMUST00000057561.9
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2 |
| chr1_-_180971763 | 0.04 |
ENSMUST00000027797.9
|
Nvl
|
nuclear VCP-like |
| chr18_+_36693646 | 0.04 |
ENSMUST00000155329.9
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr12_-_84265609 | 0.03 |
ENSMUST00000046266.13
ENSMUST00000220974.2 |
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr8_+_108669276 | 0.03 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr8_+_106245368 | 0.03 |
ENSMUST00000034363.7
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr1_-_172418058 | 0.03 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8 |
| chr15_+_98390278 | 0.03 |
ENSMUST00000205772.3
ENSMUST00000216822.2 |
Olfr279
|
olfactory receptor 279 |
| chr10_+_128139227 | 0.03 |
ENSMUST00000218315.2
ENSMUST00000219721.2 |
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr1_+_173093568 | 0.03 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr3_+_88744323 | 0.03 |
ENSMUST00000081695.14
ENSMUST00000090942.6 |
Gon4l
|
gon-4-like (C.elegans) |
| chr1_-_133728779 | 0.03 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr4_+_100633860 | 0.03 |
ENSMUST00000030257.15
ENSMUST00000097955.3 |
Cachd1
|
cache domain containing 1 |
| chr1_-_181847492 | 0.03 |
ENSMUST00000177811.8
ENSMUST00000111025.8 ENSMUST00000111024.10 |
Enah
|
ENAH actin regulator |
| chr17_-_12988492 | 0.03 |
ENSMUST00000024599.14
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr5_+_123390149 | 0.03 |
ENSMUST00000121964.8
|
Wdr66
|
WD repeat domain 66 |
| chr14_-_62998561 | 0.03 |
ENSMUST00000053959.7
ENSMUST00000223585.2 |
Ints6
|
integrator complex subunit 6 |
| chr4_-_125021610 | 0.03 |
ENSMUST00000036188.8
|
Zc3h12a
|
zinc finger CCCH type containing 12A |
| chr5_-_144294854 | 0.03 |
ENSMUST00000055190.8
|
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr7_+_107497109 | 0.03 |
ENSMUST00000209670.4
ENSMUST00000216937.3 |
Olfr472
|
olfactory receptor 472 |
| chr6_+_15720653 | 0.03 |
ENSMUST00000101663.10
ENSMUST00000190255.7 ENSMUST00000189359.7 ENSMUST00000125326.8 |
Mdfic
|
MyoD family inhibitor domain containing |
| chr8_-_106015682 | 0.03 |
ENSMUST00000212922.2
ENSMUST00000212219.2 |
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr3_+_104696108 | 0.03 |
ENSMUST00000002303.12
|
Rhoc
|
ras homolog family member C |
| chr8_-_106016097 | 0.03 |
ENSMUST00000171788.8
|
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr7_+_100757940 | 0.03 |
ENSMUST00000032931.9
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr2_+_31649946 | 0.03 |
ENSMUST00000028190.13
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr7_+_24069680 | 0.03 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr11_-_99045894 | 0.03 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr1_-_105591362 | 0.03 |
ENSMUST00000187537.7
ENSMUST00000186485.7 ENSMUST00000190811.7 |
Pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr12_+_104180795 | 0.03 |
ENSMUST00000121337.8
ENSMUST00000167049.8 ENSMUST00000101080.2 |
Serpina3f
|
serine (or cysteine) peptidase inhibitor, clade A, member 3F |
| chr10_+_58230203 | 0.03 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_-_153286361 | 0.03 |
ENSMUST00000109784.2
|
Nol4l
|
nucleolar protein 4-like |
| chr17_-_45997823 | 0.03 |
ENSMUST00000156254.8
|
Tmem63b
|
transmembrane protein 63b |
| chr2_-_166902307 | 0.03 |
ENSMUST00000155281.8
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr3_-_65865656 | 0.03 |
ENSMUST00000029416.14
|
Ccnl1
|
cyclin L1 |
| chr14_-_55828511 | 0.03 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr7_+_75498058 | 0.03 |
ENSMUST00000171155.4
ENSMUST00000092073.11 ENSMUST00000206019.2 ENSMUST00000205612.2 ENSMUST00000205887.2 |
Klhl25
|
kelch-like 25 |
| chr1_+_163875783 | 0.03 |
ENSMUST00000027874.6
|
Sele
|
selectin, endothelial cell |
| chr11_+_48977852 | 0.03 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr9_-_31043076 | 0.03 |
ENSMUST00000034478.3
|
St14
|
suppression of tumorigenicity 14 (colon carcinoma) |
| chr12_+_59113659 | 0.03 |
ENSMUST00000021381.6
|
Pnn
|
pinin |
| chr4_+_99603877 | 0.03 |
ENSMUST00000097961.9
ENSMUST00000107004.9 ENSMUST00000139799.3 |
Alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr11_+_101207743 | 0.03 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr6_+_122684448 | 0.03 |
ENSMUST00000112581.8
ENSMUST00000112580.8 ENSMUST00000012540.5 |
Nanog
|
Nanog homeobox |
| chr6_+_68233361 | 0.03 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr12_+_33364288 | 0.03 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr6_+_120750510 | 0.03 |
ENSMUST00000112682.4
|
Slc25a18
|
solute carrier family 25 (mitochondrial carrier), member 18 |
| chr2_-_109108618 | 0.03 |
ENSMUST00000081631.10
|
Mettl15
|
methyltransferase like 15 |
| chr14_-_36690636 | 0.03 |
ENSMUST00000067700.13
|
Ccser2
|
coiled-coil serine rich 2 |
| chr15_-_66158445 | 0.03 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3 |
| chr5_-_137145030 | 0.03 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr10_-_86843878 | 0.03 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr3_+_45332831 | 0.03 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr13_-_49462694 | 0.03 |
ENSMUST00000110087.9
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr10_+_79795977 | 0.03 |
ENSMUST00000045247.9
|
Wdr18
|
WD repeat domain 18 |
| chr2_+_164897547 | 0.03 |
ENSMUST00000017799.12
ENSMUST00000073707.9 |
Cd40
|
CD40 antigen |
| chr1_-_5089564 | 0.03 |
ENSMUST00000002533.15
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr16_+_13758494 | 0.03 |
ENSMUST00000141971.8
ENSMUST00000124947.8 ENSMUST00000023360.14 ENSMUST00000143697.8 |
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr18_+_37453427 | 0.03 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr12_-_69771604 | 0.03 |
ENSMUST00000021370.10
|
L2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr14_-_54655079 | 0.03 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr19_-_29625755 | 0.03 |
ENSMUST00000159692.8
|
Ermp1
|
endoplasmic reticulum metallopeptidase 1 |
| chr2_+_10158374 | 0.03 |
ENSMUST00000026886.8
|
Itih5
|
inter-alpha (globulin) inhibitor H5 |
| chr15_+_76594810 | 0.03 |
ENSMUST00000136840.8
ENSMUST00000127208.8 ENSMUST00000036423.15 ENSMUST00000137649.8 ENSMUST00000155225.2 ENSMUST00000155735.2 |
Lrrc14
|
leucine rich repeat containing 14 |
| chr14_-_70873385 | 0.03 |
ENSMUST00000228295.2
ENSMUST00000022695.16 |
Dmtn
|
dematin actin binding protein |
| chr4_+_11704438 | 0.03 |
ENSMUST00000108304.9
|
Gem
|
GTP binding protein (gene overexpressed in skeletal muscle) |
| chr9_-_105973975 | 0.02 |
ENSMUST00000121963.3
|
Col6a4
|
collagen, type VI, alpha 4 |
| chr12_+_52746158 | 0.02 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr2_+_31578537 | 0.02 |
ENSMUST00000075759.13
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr10_+_128139191 | 0.02 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr8_-_34419826 | 0.02 |
ENSMUST00000033995.14
ENSMUST00000033994.15 ENSMUST00000191473.7 ENSMUST00000053251.12 |
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr1_+_132996237 | 0.02 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr7_-_29653475 | 0.02 |
ENSMUST00000108211.8
|
Zfp74
|
zinc finger protein 74 |
| chr17_-_84154196 | 0.02 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr2_-_164285097 | 0.02 |
ENSMUST00000017153.4
|
Sdc4
|
syndecan 4 |
| chr11_-_120521382 | 0.02 |
ENSMUST00000106181.8
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr9_+_22099386 | 0.02 |
ENSMUST00000217301.2
ENSMUST00000178901.8 |
Zfp872
|
zinc finger protein 872 |
| chr3_+_40754489 | 0.02 |
ENSMUST00000203295.3
|
Plk4
|
polo like kinase 4 |
| chr5_+_90920294 | 0.02 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr7_-_100951227 | 0.02 |
ENSMUST00000122116.8
ENSMUST00000120267.9 |
Atg16l2
|
autophagy related 16-like 2 (S. cerevisiae) |
| chr12_+_111132908 | 0.02 |
ENSMUST00000139162.8
ENSMUST00000060274.7 |
Traf3
|
TNF receptor-associated factor 3 |
| chr7_-_5128936 | 0.02 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr6_-_127086480 | 0.02 |
ENSMUST00000039913.9
|
Tigar
|
Trp53 induced glycolysis regulatory phosphatase |
| chr2_+_92015780 | 0.02 |
ENSMUST00000128781.9
ENSMUST00000111291.9 |
Phf21a
|
PHD finger protein 21A |
| chr7_-_43155351 | 0.02 |
ENSMUST00000191516.7
ENSMUST00000013497.8 ENSMUST00000163619.8 |
4931406B18Rik
|
RIKEN cDNA 4931406B18 gene |
| chr5_-_135518098 | 0.02 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr11_-_84761637 | 0.02 |
ENSMUST00000168434.8
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr17_-_34043320 | 0.02 |
ENSMUST00000173879.8
ENSMUST00000166693.3 ENSMUST00000173019.8 |
Rps28
|
ribosomal protein S28 |
| chr3_+_115681788 | 0.02 |
ENSMUST00000196804.5
ENSMUST00000106505.8 ENSMUST00000043342.10 |
Dph5
|
diphthamide biosynthesis 5 |
| chr6_-_83418656 | 0.02 |
ENSMUST00000089622.11
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr11_+_94900677 | 0.02 |
ENSMUST00000055947.10
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr6_-_24664959 | 0.02 |
ENSMUST00000041737.8
ENSMUST00000031695.15 |
Wasl
|
WASP like actin nucleation promoting factor |
| chr5_+_137020478 | 0.02 |
ENSMUST00000127100.2
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr1_-_75168325 | 0.02 |
ENSMUST00000186744.2
|
Atg9a
|
autophagy related 9A |
| chr7_-_87142580 | 0.02 |
ENSMUST00000207834.2
ENSMUST00000004770.7 |
Tyr
|
tyrosinase |
| chr17_+_37581103 | 0.02 |
ENSMUST00000038580.7
|
H2-M3
|
histocompatibility 2, M region locus 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rela_Rel_Nfkb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.1 | 0.4 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.0 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.0 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.0 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.0 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0033643 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0004371 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |