Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
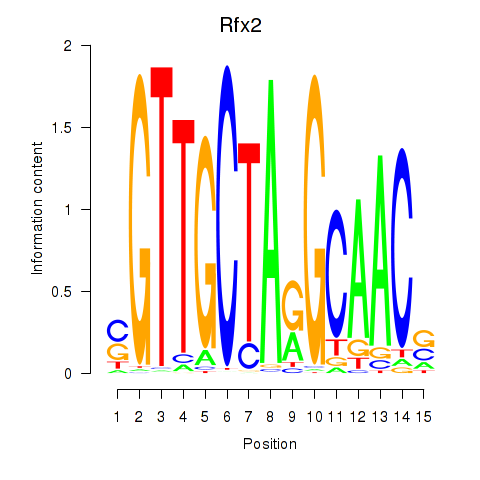
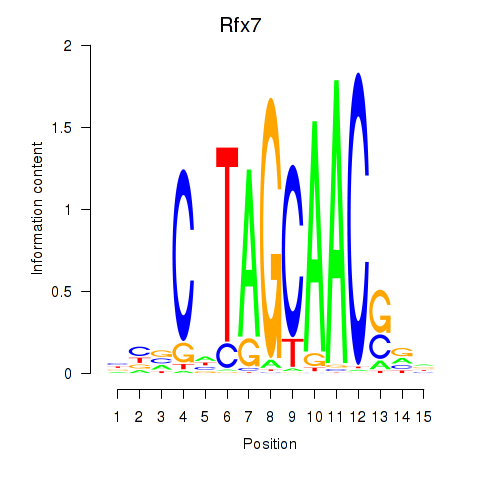
Results for Rfx2_Rfx7
Z-value: 1.53
Transcription factors associated with Rfx2_Rfx7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx2
|
ENSMUSG00000024206.16 | regulatory factor X, 2 (influences HLA class II expression) |
|
Rfx7
|
ENSMUSG00000037674.16 | regulatory factor X, 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx2 | mm39_v1_chr17_-_57137898_57138013 | -0.96 | 9.9e-03 | Click! |
| Rfx7 | mm39_v1_chr9_+_72439496_72439553 | -0.58 | 3.1e-01 | Click! |
Activity profile of Rfx2_Rfx7 motif
Sorted Z-values of Rfx2_Rfx7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_39472981 | 1.34 |
ENSMUST00000239508.1
ENSMUST00000239509.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr8_+_72994152 | 1.22 |
ENSMUST00000126885.2
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr3_-_50398027 | 1.12 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr2_+_21210527 | 0.94 |
ENSMUST00000054591.10
ENSMUST00000102952.8 ENSMUST00000138965.8 ENSMUST00000138914.8 ENSMUST00000102951.2 |
Thnsl1
|
threonine synthase-like 1 (bacterial) |
| chr19_-_10859046 | 0.88 |
ENSMUST00000128835.8
|
Tmem109
|
transmembrane protein 109 |
| chr7_+_44117511 | 0.85 |
ENSMUST00000121922.3
ENSMUST00000208117.2 |
Josd2
|
Josephin domain containing 2 |
| chr7_-_4687916 | 0.84 |
ENSMUST00000206306.2
ENSMUST00000205952.2 ENSMUST00000079970.6 |
Hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr5_-_117527094 | 0.75 |
ENSMUST00000111953.2
ENSMUST00000086461.13 |
Rfc5
|
replication factor C (activator 1) 5 |
| chr7_+_44117444 | 0.72 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr2_+_80145805 | 0.69 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr7_-_144493560 | 0.68 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr2_+_71219561 | 0.64 |
ENSMUST00000028408.3
|
Hat1
|
histone aminotransferase 1 |
| chrX_+_134786600 | 0.59 |
ENSMUST00000180025.8
ENSMUST00000148374.8 ENSMUST00000068755.14 |
Bhlhb9
|
basic helix-loop-helix domain containing, class B9 |
| chr7_+_44117475 | 0.59 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr11_+_59839032 | 0.54 |
ENSMUST00000081980.7
|
Med9
|
mediator complex subunit 9 |
| chr15_-_81810349 | 0.52 |
ENSMUST00000023113.7
|
Polr3h
|
polymerase (RNA) III (DNA directed) polypeptide H |
| chr6_+_83162928 | 0.52 |
ENSMUST00000113907.2
|
Dctn1
|
dynactin 1 |
| chr2_+_32665781 | 0.50 |
ENSMUST00000066352.6
|
Ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr2_-_28730286 | 0.50 |
ENSMUST00000037117.6
ENSMUST00000171404.8 |
Gtf3c4
|
general transcription factor IIIC, polypeptide 4 |
| chr4_-_12087911 | 0.49 |
ENSMUST00000050686.10
|
Tmem67
|
transmembrane protein 67 |
| chr2_-_125701059 | 0.49 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr14_-_56339915 | 0.46 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr3_+_108164242 | 0.46 |
ENSMUST00000090569.10
|
Psma5
|
proteasome subunit alpha 5 |
| chr11_+_49135018 | 0.46 |
ENSMUST00000167400.8
ENSMUST00000081794.7 |
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr1_-_75187417 | 0.45 |
ENSMUST00000113623.8
|
Glb1l
|
galactosidase, beta 1-like |
| chr2_+_125701054 | 0.45 |
ENSMUST00000028636.13
ENSMUST00000125084.8 |
Galk2
|
galactokinase 2 |
| chr15_-_79967543 | 0.44 |
ENSMUST00000081650.15
|
Rpl3
|
ribosomal protein L3 |
| chr11_-_102771751 | 0.43 |
ENSMUST00000021306.14
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr1_+_132433968 | 0.43 |
ENSMUST00000058167.3
|
Tmem81
|
transmembrane protein 81 |
| chr13_-_55684317 | 0.43 |
ENSMUST00000021956.9
ENSMUST00000224765.2 |
Ddx41
|
DEAD box helicase 41 |
| chr7_-_79882313 | 0.42 |
ENSMUST00000206084.2
ENSMUST00000205996.2 ENSMUST00000071457.12 |
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chrX_+_93410718 | 0.42 |
ENSMUST00000113898.8
|
Apoo
|
apolipoprotein O |
| chr7_-_99508117 | 0.42 |
ENSMUST00000209032.2
ENSMUST00000036274.8 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr2_-_125700905 | 0.41 |
ENSMUST00000110462.8
|
Cops2
|
COP9 signalosome subunit 2 |
| chr7_+_44117404 | 0.41 |
ENSMUST00000035844.11
|
Josd2
|
Josephin domain containing 2 |
| chr11_-_102771806 | 0.40 |
ENSMUST00000107060.8
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr3_+_146110387 | 0.40 |
ENSMUST00000106151.8
ENSMUST00000106153.9 ENSMUST00000039021.11 ENSMUST00000106149.8 ENSMUST00000149262.8 |
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr17_-_87573294 | 0.40 |
ENSMUST00000145895.8
ENSMUST00000129616.8 ENSMUST00000155904.2 ENSMUST00000151155.8 ENSMUST00000144236.9 ENSMUST00000024963.11 |
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr17_+_8502682 | 0.39 |
ENSMUST00000124023.8
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr4_-_133694607 | 0.39 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr10_+_111309020 | 0.38 |
ENSMUST00000065917.16
ENSMUST00000219961.2 ENSMUST00000217908.2 |
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chrX_+_93410754 | 0.38 |
ENSMUST00000113897.9
ENSMUST00000113896.8 ENSMUST00000113895.2 |
Apoo
|
apolipoprotein O |
| chr1_+_191553556 | 0.37 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr7_-_99508066 | 0.36 |
ENSMUST00000208477.2
ENSMUST00000208465.2 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr5_-_137529251 | 0.36 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr7_-_125090540 | 0.36 |
ENSMUST00000138616.3
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr9_-_108455899 | 0.35 |
ENSMUST00000068700.7
|
Wdr6
|
WD repeat domain 6 |
| chr17_-_30795403 | 0.34 |
ENSMUST00000237037.2
ENSMUST00000168787.8 |
Btbd9
|
BTB (POZ) domain containing 9 |
| chr17_-_46558894 | 0.34 |
ENSMUST00000142706.9
ENSMUST00000173349.8 ENSMUST00000087026.13 |
Polr1c
|
polymerase (RNA) I polypeptide C |
| chr4_-_152112087 | 0.33 |
ENSMUST00000131935.2
ENSMUST00000066715.11 |
Zbtb48
|
zinc finger and BTB domain containing 48 |
| chr10_-_85752765 | 0.33 |
ENSMUST00000037646.9
ENSMUST00000220032.2 |
Prdm4
|
PR domain containing 4 |
| chr2_-_152239966 | 0.33 |
ENSMUST00000063332.9
ENSMUST00000182625.2 |
Sox12
|
SRY (sex determining region Y)-box 12 |
| chr2_+_30331839 | 0.32 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr8_+_110944575 | 0.32 |
ENSMUST00000056972.6
|
Cmtr2
|
cap methyltransferase 2 |
| chr8_+_34621717 | 0.32 |
ENSMUST00000239436.2
ENSMUST00000033933.8 |
Saraf
|
store-operated calcium entry-associated regulatory factor |
| chr11_+_97697328 | 0.32 |
ENSMUST00000153520.3
|
Lasp1
|
LIM and SH3 protein 1 |
| chr4_-_123644091 | 0.32 |
ENSMUST00000102636.4
|
Akirin1
|
akirin 1 |
| chr13_-_74465353 | 0.31 |
ENSMUST00000022060.7
|
Pdcd6
|
programmed cell death 6 |
| chr7_-_79882501 | 0.31 |
ENSMUST00000065163.15
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr12_+_111132779 | 0.31 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3 |
| chr5_-_41865461 | 0.31 |
ENSMUST00000201422.4
|
Rab28
|
RAB28, member RAS oncogene family |
| chr2_+_125994050 | 0.31 |
ENSMUST00000170908.8
|
Dtwd1
|
DTW domain containing 1 |
| chr5_-_41865477 | 0.30 |
ENSMUST00000031011.12
ENSMUST00000202913.2 |
Rab28
|
RAB28, member RAS oncogene family |
| chr7_+_100145192 | 0.30 |
ENSMUST00000133044.3
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_-_119190830 | 0.30 |
ENSMUST00000106253.2
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr16_-_91525485 | 0.30 |
ENSMUST00000231499.2
ENSMUST00000141664.9 ENSMUST00000123751.2 ENSMUST00000122254.8 ENSMUST00000114023.3 |
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr11_-_23583591 | 0.30 |
ENSMUST00000180260.8
ENSMUST00000141353.8 ENSMUST00000131612.2 ENSMUST00000109532.9 |
0610010F05Rik
|
RIKEN cDNA 0610010F05 gene |
| chr14_-_30723549 | 0.29 |
ENSMUST00000226782.2
ENSMUST00000186131.7 ENSMUST00000228767.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr1_+_75187474 | 0.29 |
ENSMUST00000027401.11
ENSMUST00000144355.8 ENSMUST00000123825.8 ENSMUST00000189698.7 |
Stk16
|
serine/threonine kinase 16 |
| chr13_+_120151982 | 0.29 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr14_-_31362835 | 0.29 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr7_+_46496552 | 0.28 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr19_+_8779903 | 0.28 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr7_+_46496506 | 0.28 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr3_+_37366662 | 0.28 |
ENSMUST00000138710.3
ENSMUST00000057975.8 ENSMUST00000108121.4 |
Gm43439
Bbs12
|
predicted gene 43439 Bardet-Biedl syndrome 12 (human) |
| chr7_+_18910340 | 0.28 |
ENSMUST00000117338.8
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr3_-_107838895 | 0.28 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr11_+_96932379 | 0.28 |
ENSMUST00000001485.10
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr8_-_106434565 | 0.28 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr7_+_46496929 | 0.28 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr18_-_24153363 | 0.27 |
ENSMUST00000153337.2
ENSMUST00000148525.2 |
Zfp24
|
zinc finger protein 24 |
| chr12_+_33004178 | 0.27 |
ENSMUST00000020885.13
|
Sypl
|
synaptophysin-like protein |
| chr15_-_79718423 | 0.27 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr10_-_90959817 | 0.27 |
ENSMUST00000164505.2
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr2_+_28730418 | 0.27 |
ENSMUST00000113853.3
|
Ddx31
|
DEAD/H box helicase 31 |
| chr7_+_3632982 | 0.26 |
ENSMUST00000179769.8
ENSMUST00000008517.13 |
Prpf31
|
pre-mRNA processing factor 31 |
| chr10_-_22607136 | 0.26 |
ENSMUST00000238910.2
ENSMUST00000127698.8 |
ENSMUSG00000118528.2
Tbpl1
|
novel protein TATA box binding protein-like 1 |
| chr19_-_38032006 | 0.26 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr5_-_137529465 | 0.26 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr2_-_164876690 | 0.26 |
ENSMUST00000122070.2
ENSMUST00000121377.8 ENSMUST00000153905.2 ENSMUST00000040381.15 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr7_-_125090757 | 0.26 |
ENSMUST00000033006.14
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr1_-_131066004 | 0.26 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr6_+_89620956 | 0.25 |
ENSMUST00000000828.14
ENSMUST00000101171.3 |
Txnrd3
|
thioredoxin reductase 3 |
| chr16_-_91525655 | 0.25 |
ENSMUST00000117644.8
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr17_+_35827997 | 0.25 |
ENSMUST00000164242.9
ENSMUST00000045956.14 |
Cchcr1
|
coiled-coil alpha-helical rod protein 1 |
| chrX_-_71962017 | 0.25 |
ENSMUST00000114551.10
|
Cetn2
|
centrin 2 |
| chr9_-_21061196 | 0.25 |
ENSMUST00000215296.2
ENSMUST00000019615.11 |
Cdc37
|
cell division cycle 37 |
| chr7_+_26932425 | 0.24 |
ENSMUST00000003860.13
ENSMUST00000108378.10 |
Coq8b
|
coenzyme Q8B |
| chr7_+_127187910 | 0.24 |
ENSMUST00000205694.2
ENSMUST00000033088.8 ENSMUST00000206914.2 |
Rnf40
|
ring finger protein 40 |
| chr11_-_59937302 | 0.24 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr12_+_76812301 | 0.24 |
ENSMUST00000041262.14
ENSMUST00000126408.2 ENSMUST00000110399.3 ENSMUST00000137826.8 |
Churc1
Fntb
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta |
| chr16_-_91525863 | 0.24 |
ENSMUST00000073466.13
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr17_-_26080429 | 0.24 |
ENSMUST00000079461.15
ENSMUST00000176923.9 |
Wdr90
|
WD repeat domain 90 |
| chr7_-_79882228 | 0.24 |
ENSMUST00000123279.8
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr5_+_24305577 | 0.24 |
ENSMUST00000030841.10
ENSMUST00000163409.5 |
Klhl7
|
kelch-like 7 |
| chr2_-_65955338 | 0.23 |
ENSMUST00000028378.4
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr19_-_40600619 | 0.23 |
ENSMUST00000132452.2
ENSMUST00000135795.8 ENSMUST00000025981.15 |
Tctn3
|
tectonic family member 3 |
| chr12_+_70499869 | 0.23 |
ENSMUST00000021471.13
|
Tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr4_-_132649798 | 0.23 |
ENSMUST00000097856.10
ENSMUST00000030696.11 |
Fam76a
|
family with sequence similarity 76, member A |
| chr1_-_75187441 | 0.23 |
ENSMUST00000185448.2
|
Glb1l
|
galactosidase, beta 1-like |
| chr7_-_16761732 | 0.22 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr18_-_24153805 | 0.22 |
ENSMUST00000066497.12
|
Zfp24
|
zinc finger protein 24 |
| chr12_+_113120023 | 0.22 |
ENSMUST00000049271.13
|
Tedc1
|
tubulin epsilon and delta complex 1 |
| chr13_-_100912308 | 0.22 |
ENSMUST00000075550.4
|
Cenph
|
centromere protein H |
| chr7_-_143153785 | 0.22 |
ENSMUST00000105909.4
ENSMUST00000010899.14 |
Cars
|
cysteinyl-tRNA synthetase |
| chr11_-_119190896 | 0.21 |
ENSMUST00000026667.15
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr2_+_162916551 | 0.21 |
ENSMUST00000142729.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr11_+_117006020 | 0.21 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chr12_+_111132847 | 0.21 |
ENSMUST00000021706.11
|
Traf3
|
TNF receptor-associated factor 3 |
| chr7_+_66339637 | 0.21 |
ENSMUST00000153007.2
ENSMUST00000121777.9 ENSMUST00000150071.8 ENSMUST00000077967.13 |
Lins1
|
lines homolog 1 |
| chr4_-_133694543 | 0.21 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_119833589 | 0.21 |
ENSMUST00000106231.8
ENSMUST00000075180.12 ENSMUST00000103021.10 ENSMUST00000026436.10 ENSMUST00000106233.2 |
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2 |
| chr10_+_111309127 | 0.21 |
ENSMUST00000219143.2
|
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr4_+_128582519 | 0.20 |
ENSMUST00000106080.8
|
Phc2
|
polyhomeotic 2 |
| chr14_-_30723292 | 0.20 |
ENSMUST00000228736.2
ENSMUST00000226374.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr12_+_33003882 | 0.20 |
ENSMUST00000076698.13
|
Sypl
|
synaptophysin-like protein |
| chr2_-_152673585 | 0.20 |
ENSMUST00000156688.2
ENSMUST00000007803.12 |
Bcl2l1
|
BCL2-like 1 |
| chr19_-_8858247 | 0.20 |
ENSMUST00000096253.7
|
Uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr4_+_137989783 | 0.20 |
ENSMUST00000105821.3
|
Kif17
|
kinesin family member 17 |
| chr8_-_84059048 | 0.20 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr14_+_55909816 | 0.19 |
ENSMUST00000227178.2
ENSMUST00000227914.2 |
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr12_+_98886826 | 0.19 |
ENSMUST00000085109.10
ENSMUST00000079146.13 |
Ttc8
|
tetratricopeptide repeat domain 8 |
| chr1_+_63769772 | 0.19 |
ENSMUST00000027103.7
|
Fastkd2
|
FAST kinase domains 2 |
| chr12_-_40088024 | 0.19 |
ENSMUST00000101472.4
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr5_+_21850975 | 0.19 |
ENSMUST00000095495.3
|
Armc10
|
armadillo repeat containing 10 |
| chr9_-_21202353 | 0.19 |
ENSMUST00000086374.8
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr7_-_80550968 | 0.18 |
ENSMUST00000146402.2
ENSMUST00000026816.15 |
Wdr73
|
WD repeat domain 73 |
| chr2_+_172841907 | 0.18 |
ENSMUST00000029013.10
ENSMUST00000132212.2 |
Rae1
|
ribonucleic acid export 1 |
| chr8_-_85751897 | 0.18 |
ENSMUST00000064314.10
|
Get3
|
guided entry of tail-anchored proteins factor 3, ATPase |
| chr13_+_83652352 | 0.18 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr8_-_65186565 | 0.18 |
ENSMUST00000141021.2
|
Msmo1
|
methylsterol monoxygenase 1 |
| chr5_+_135807334 | 0.18 |
ENSMUST00000019323.11
|
Mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr14_+_55909692 | 0.18 |
ENSMUST00000002397.7
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr12_-_85335193 | 0.18 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr1_-_36586275 | 0.18 |
ENSMUST00000194894.4
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr5_+_130173702 | 0.18 |
ENSMUST00000040616.9
|
Kctd7
|
potassium channel tetramerisation domain containing 7 |
| chr18_+_31742565 | 0.18 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chrX_+_133587268 | 0.17 |
ENSMUST00000124226.3
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr4_+_118266582 | 0.17 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr12_-_87247082 | 0.17 |
ENSMUST00000037418.7
|
Tmed8
|
transmembrane p24 trafficking protein 8 |
| chr19_+_29078765 | 0.17 |
ENSMUST00000064393.6
ENSMUST00000235900.2 |
Rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr13_+_12410240 | 0.17 |
ENSMUST00000059270.10
|
Heatr1
|
HEAT repeat containing 1 |
| chr17_+_43878989 | 0.17 |
ENSMUST00000167214.8
ENSMUST00000024706.12 |
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr1_-_59276252 | 0.17 |
ENSMUST00000163058.2
ENSMUST00000160945.2 ENSMUST00000027178.13 |
Als2
|
alsin Rho guanine nucleotide exchange factor |
| chr2_+_145745154 | 0.17 |
ENSMUST00000110000.8
ENSMUST00000002805.14 ENSMUST00000169732.8 ENSMUST00000134759.3 |
Naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr19_-_43512929 | 0.17 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr17_+_8502594 | 0.17 |
ENSMUST00000155364.8
ENSMUST00000046754.15 |
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr17_+_32255071 | 0.17 |
ENSMUST00000081339.13
|
Rrp1b
|
ribosomal RNA processing 1B |
| chrX_-_7999009 | 0.17 |
ENSMUST00000130832.8
ENSMUST00000033506.13 ENSMUST00000115623.8 ENSMUST00000153839.2 |
Wdr13
|
WD repeat domain 13 |
| chr2_+_30171055 | 0.17 |
ENSMUST00000143119.3
|
Gm28038
|
predicted gene, 28038 |
| chr3_+_138148846 | 0.16 |
ENSMUST00000005964.7
|
Adh5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr14_-_31362909 | 0.16 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr15_-_89361571 | 0.16 |
ENSMUST00000165199.8
|
Arsa
|
arylsulfatase A |
| chr4_-_44084167 | 0.16 |
ENSMUST00000030201.14
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr7_-_55669702 | 0.16 |
ENSMUST00000052204.6
|
Nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) |
| chr5_-_149559636 | 0.16 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr12_+_78908466 | 0.16 |
ENSMUST00000071230.8
|
Eif2s1
|
eukaryotic translation initiation factor 2, subunit 1 alpha |
| chr15_+_79113341 | 0.16 |
ENSMUST00000163571.8
|
Pick1
|
protein interacting with C kinase 1 |
| chr13_+_97377604 | 0.15 |
ENSMUST00000041623.9
|
Enc1
|
ectodermal-neural cortex 1 |
| chr15_+_78312764 | 0.15 |
ENSMUST00000162517.8
ENSMUST00000166142.10 ENSMUST00000089414.11 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr16_+_36832119 | 0.15 |
ENSMUST00000071452.12
ENSMUST00000054034.7 |
Polq
|
polymerase (DNA directed), theta |
| chr5_-_149559667 | 0.15 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_-_121495623 | 0.15 |
ENSMUST00000001724.12
|
Ddx18
|
DEAD box helicase 18 |
| chr1_-_52271455 | 0.15 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr10_+_128247598 | 0.15 |
ENSMUST00000096386.13
|
Rnf41
|
ring finger protein 41 |
| chr14_+_31363004 | 0.15 |
ENSMUST00000090147.7
|
Btd
|
biotinidase |
| chr2_+_164587948 | 0.15 |
ENSMUST00000109327.4
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr3_-_144275897 | 0.15 |
ENSMUST00000043325.9
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr9_-_21202693 | 0.15 |
ENSMUST00000213407.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr9_-_106563091 | 0.15 |
ENSMUST00000046735.11
|
Tex264
|
testis expressed gene 264 |
| chrX_+_163763588 | 0.15 |
ENSMUST00000167446.8
ENSMUST00000057150.8 |
Fancb
|
Fanconi anemia, complementation group B |
| chr3_+_88483183 | 0.15 |
ENSMUST00000192688.6
ENSMUST00000193069.2 ENSMUST00000194604.2 |
Ssr2
|
signal sequence receptor, beta |
| chr11_+_96932395 | 0.15 |
ENSMUST00000054252.5
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr15_+_78798116 | 0.15 |
ENSMUST00000089378.5
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr8_+_112667328 | 0.14 |
ENSMUST00000034428.8
|
Gabarapl2
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 2 |
| chr15_-_54953819 | 0.14 |
ENSMUST00000110231.2
ENSMUST00000023059.13 |
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr3_+_144824325 | 0.14 |
ENSMUST00000098538.9
ENSMUST00000106192.9 ENSMUST00000098539.7 ENSMUST00000029920.15 |
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr2_-_152673032 | 0.14 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr9_+_63509925 | 0.14 |
ENSMUST00000041551.9
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr6_+_67838100 | 0.14 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr11_-_116472272 | 0.14 |
ENSMUST00000082152.5
|
Ube2o
|
ubiquitin-conjugating enzyme E2O |
| chr11_+_117005958 | 0.14 |
ENSMUST00000021177.15
|
Sec14l1
|
SEC14-like lipid binding 1 |
| chr19_-_16758268 | 0.14 |
ENSMUST00000068156.8
|
Vps13a
|
vacuolar protein sorting 13A |
| chr6_+_8259379 | 0.14 |
ENSMUST00000162034.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr11_+_17161912 | 0.14 |
ENSMUST00000046955.7
|
Wdr92
|
WD repeat domain 92 |
| chr4_-_72770859 | 0.14 |
ENSMUST00000179234.2
ENSMUST00000078617.5 |
Aldoart1
|
aldolase 1 A, retrogene 1 |
| chr2_-_126333450 | 0.14 |
ENSMUST00000040149.13
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr2_+_166634445 | 0.14 |
ENSMUST00000065753.2
|
Trp53rkb
|
transformation related protein 53 regulating kinase B |
| chr13_+_21363602 | 0.14 |
ENSMUST00000222544.2
|
Trim27
|
tripartite motif-containing 27 |
| chr3_+_144276154 | 0.13 |
ENSMUST00000082437.11
|
Selenof
|
selenoprotein F |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx2_Rfx7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.2 | 0.5 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.5 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.4 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.1 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.3 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 1.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.7 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.8 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 0.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 1.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.3 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.1 | 0.2 | GO:0006532 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.3 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.3 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.2 | GO:0046294 | ethanol oxidation(GO:0006069) formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.3 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.3 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.1 | 0.5 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.5 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.5 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.2 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.4 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.5 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.4 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transport(GO:2000680) regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 1.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0060287 | determination of pancreatic left/right asymmetry(GO:0035469) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 1.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 1.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.3 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.3 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.7 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.7 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.2 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 1.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0016297 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-acetyltransferase activity(GO:0016418) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 1.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |