Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
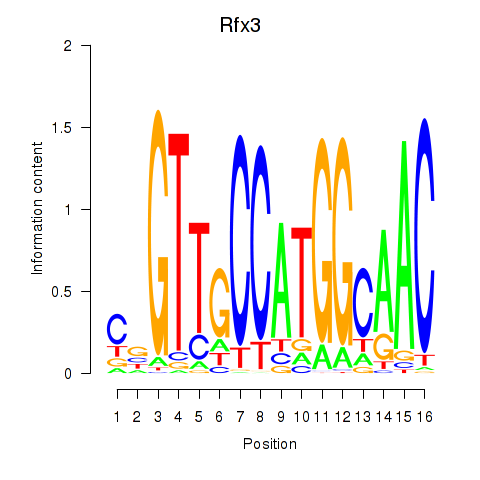
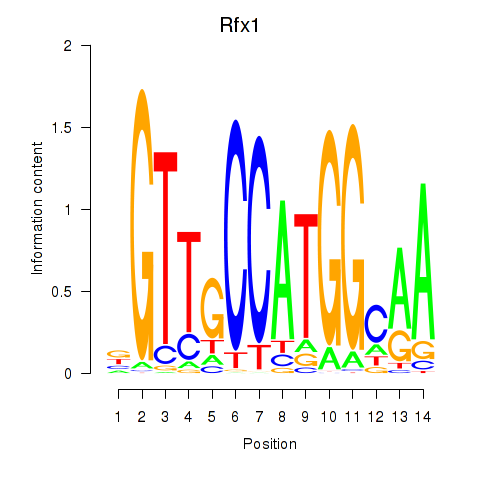
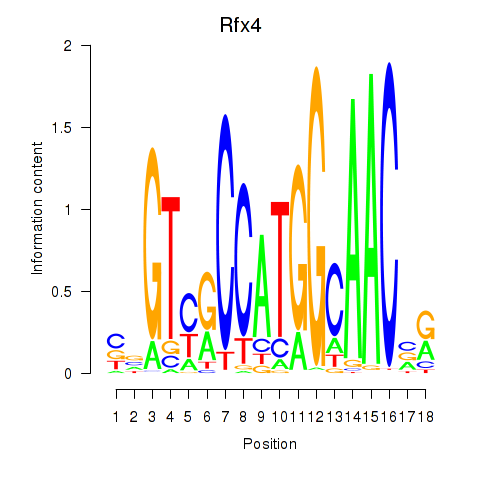
Results for Rfx3_Rfx1_Rfx4
Z-value: 1.31
Transcription factors associated with Rfx3_Rfx1_Rfx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx3
|
ENSMUSG00000040929.18 | regulatory factor X, 3 (influences HLA class II expression) |
|
Rfx1
|
ENSMUSG00000031706.8 | regulatory factor X, 1 (influences HLA class II expression) |
|
Rfx4
|
ENSMUSG00000020037.16 | regulatory factor X, 4 (influences HLA class II expression) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx4 | mm39_v1_chr10_+_84674008_84674016 | -0.83 | 8.0e-02 | Click! |
| Rfx1 | mm39_v1_chr8_+_84793453_84793511 | -0.77 | 1.3e-01 | Click! |
| Rfx3 | mm39_v1_chr19_-_27988534_27988581 | -0.76 | 1.3e-01 | Click! |
Activity profile of Rfx3_Rfx1_Rfx4 motif
Sorted Z-values of Rfx3_Rfx1_Rfx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_43031370 | 1.52 |
ENSMUST00000138030.2
ENSMUST00000136326.8 |
Stoml2
|
stomatin (Epb7.2)-like 2 |
| chr11_+_87651359 | 0.86 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr4_-_94940425 | 0.84 |
ENSMUST00000107094.2
|
Jun
|
jun proto-oncogene |
| chr5_-_137529251 | 0.63 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr10_+_61431271 | 0.62 |
ENSMUST00000020287.8
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr5_-_137074880 | 0.61 |
ENSMUST00000144303.2
ENSMUST00000111080.8 |
Ap1s1
|
adaptor protein complex AP-1, sigma 1 |
| chr17_-_35055843 | 0.60 |
ENSMUST00000077477.12
|
Stk19
|
serine/threonine kinase 19 |
| chr2_-_91274967 | 0.55 |
ENSMUST00000064652.14
ENSMUST00000102594.11 ENSMUST00000094835.9 |
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene |
| chr15_-_78947038 | 0.53 |
ENSMUST00000151889.8
ENSMUST00000040676.11 |
Ankrd54
|
ankyrin repeat domain 54 |
| chr14_+_51193449 | 0.52 |
ENSMUST00000095925.5
|
Pnp2
|
purine-nucleoside phosphorylase 2 |
| chr4_-_133694607 | 0.52 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr2_+_80145805 | 0.52 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr9_+_107926502 | 0.47 |
ENSMUST00000047947.9
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr5_-_137529465 | 0.47 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr7_+_18910340 | 0.45 |
ENSMUST00000117338.8
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr4_-_129556234 | 0.45 |
ENSMUST00000003828.11
|
Kpna6
|
karyopherin (importin) alpha 6 |
| chr7_+_18883647 | 0.44 |
ENSMUST00000049294.4
|
Snrpd2
|
small nuclear ribonucleoprotein D2 |
| chr6_+_88061464 | 0.43 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr3_-_146476331 | 0.43 |
ENSMUST00000106138.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr2_+_29678248 | 0.43 |
ENSMUST00000028137.10
ENSMUST00000148791.2 |
Coq4
|
coenzyme Q4 |
| chr8_-_123939480 | 0.43 |
ENSMUST00000000759.9
|
Chmp1a
|
charged multivesicular body protein 1A |
| chr3_-_146475974 | 0.42 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr17_-_46558894 | 0.42 |
ENSMUST00000142706.9
ENSMUST00000173349.8 ENSMUST00000087026.13 |
Polr1c
|
polymerase (RNA) I polypeptide C |
| chr3_+_103875574 | 0.42 |
ENSMUST00000063717.14
ENSMUST00000055425.15 ENSMUST00000123611.8 ENSMUST00000090685.11 |
Phtf1
|
putative homeodomain transcription factor 1 |
| chr9_-_60594742 | 0.42 |
ENSMUST00000114032.8
ENSMUST00000166168.8 ENSMUST00000132366.2 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr2_-_29938841 | 0.41 |
ENSMUST00000113711.3
|
Dync2i2
|
dynein 2 intermediate chain 2 |
| chr17_-_80514725 | 0.40 |
ENSMUST00000234696.2
ENSMUST00000235069.2 ENSMUST00000063417.11 |
Srsf7
|
serine and arginine-rich splicing factor 7 |
| chr5_+_137756407 | 0.40 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr10_-_116899664 | 0.40 |
ENSMUST00000218719.2
ENSMUST00000219573.2 ENSMUST00000047672.9 |
Cct2
|
chaperonin containing Tcp1, subunit 2 (beta) |
| chr7_-_4687916 | 0.40 |
ENSMUST00000206306.2
ENSMUST00000205952.2 ENSMUST00000079970.6 |
Hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr7_-_45480200 | 0.40 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr8_-_65186565 | 0.40 |
ENSMUST00000141021.2
|
Msmo1
|
methylsterol monoxygenase 1 |
| chr15_+_79543397 | 0.40 |
ENSMUST00000023064.9
|
Cby1
|
chibby family member 1, beta catenin antagonist |
| chr17_+_35055962 | 0.39 |
ENSMUST00000173874.8
ENSMUST00000180043.8 ENSMUST00000046244.15 |
Dxo
|
decapping exoribonuclease |
| chr2_-_127630769 | 0.39 |
ENSMUST00000028857.14
ENSMUST00000110357.2 |
Nphp1
|
nephronophthisis 1 (juvenile) homolog (human) |
| chr12_+_82216193 | 0.38 |
ENSMUST00000166429.9
ENSMUST00000220963.2 |
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr19_+_45549009 | 0.38 |
ENSMUST00000047057.9
|
Gm17018
|
predicted gene 17018 |
| chr5_-_115438971 | 0.38 |
ENSMUST00000112090.2
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr5_-_41865461 | 0.38 |
ENSMUST00000201422.4
|
Rab28
|
RAB28, member RAS oncogene family |
| chr1_-_156501860 | 0.38 |
ENSMUST00000188964.7
ENSMUST00000190607.2 ENSMUST00000079625.11 |
Tor3a
|
torsin family 3, member A |
| chr11_-_119190830 | 0.37 |
ENSMUST00000106253.2
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr14_+_51181956 | 0.37 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr5_-_41865477 | 0.37 |
ENSMUST00000031011.12
ENSMUST00000202913.2 |
Rab28
|
RAB28, member RAS oncogene family |
| chr10_+_80905869 | 0.36 |
ENSMUST00000005057.7
|
Thop1
|
thimet oligopeptidase 1 |
| chr17_-_26014613 | 0.36 |
ENSMUST00000235889.2
|
Gm50367
|
predicted gene, 50367 |
| chr9_-_60595401 | 0.36 |
ENSMUST00000114034.9
ENSMUST00000065603.12 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr7_+_79836581 | 0.36 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr12_-_83609217 | 0.36 |
ENSMUST00000222448.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr16_-_91525485 | 0.35 |
ENSMUST00000231499.2
ENSMUST00000141664.9 ENSMUST00000123751.2 ENSMUST00000122254.8 ENSMUST00000114023.3 |
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr15_+_81548090 | 0.35 |
ENSMUST00000023029.15
ENSMUST00000174229.8 ENSMUST00000172748.8 |
L3mbtl2
|
L3MBTL2 polycomb repressive complex 1 subunit |
| chr12_+_84408803 | 0.34 |
ENSMUST00000110278.8
ENSMUST00000145522.2 |
Coq6
|
coenzyme Q6 monooxygenase |
| chr7_-_12788592 | 0.34 |
ENSMUST00000182515.8
ENSMUST00000069289.15 |
Mzf1
|
myeloid zinc finger 1 |
| chr1_-_192946359 | 0.34 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr14_-_30723549 | 0.33 |
ENSMUST00000226782.2
ENSMUST00000186131.7 ENSMUST00000228767.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr7_-_12788441 | 0.33 |
ENSMUST00000182087.2
|
Mzf1
|
myeloid zinc finger 1 |
| chr7_-_118304930 | 0.33 |
ENSMUST00000207323.2
ENSMUST00000038791.15 |
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr12_+_84408742 | 0.33 |
ENSMUST00000021661.13
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr2_+_21210527 | 0.32 |
ENSMUST00000054591.10
ENSMUST00000102952.8 ENSMUST00000138965.8 ENSMUST00000138914.8 ENSMUST00000102951.2 |
Thnsl1
|
threonine synthase-like 1 (bacterial) |
| chr19_-_45548942 | 0.32 |
ENSMUST00000026239.7
|
Poll
|
polymerase (DNA directed), lambda |
| chr17_+_26048010 | 0.32 |
ENSMUST00000026832.14
|
Jmjd8
|
jumonji domain containing 8 |
| chr2_-_90735171 | 0.32 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr5_-_124616393 | 0.32 |
ENSMUST00000031347.8
|
Rilpl2
|
Rab interacting lysosomal protein-like 2 |
| chr10_-_79710067 | 0.31 |
ENSMUST00000166023.2
ENSMUST00000167707.2 ENSMUST00000165601.8 |
Plppr3
|
phospholipid phosphatase related 3 |
| chr15_+_100659622 | 0.31 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr12_-_86773160 | 0.31 |
ENSMUST00000021682.9
|
Angel1
|
angel homolog 1 |
| chr6_+_83162928 | 0.31 |
ENSMUST00000113907.2
|
Dctn1
|
dynactin 1 |
| chr1_+_191553556 | 0.31 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr2_-_134486039 | 0.31 |
ENSMUST00000038228.11
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr11_+_68961599 | 0.30 |
ENSMUST00000075980.12
ENSMUST00000094081.5 |
Tmem107
|
transmembrane protein 107 |
| chr19_-_10533088 | 0.30 |
ENSMUST00000059582.9
ENSMUST00000154383.2 |
Tmem216
|
transmembrane protein 216 |
| chr11_-_119190896 | 0.30 |
ENSMUST00000026667.15
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr7_+_140462343 | 0.30 |
ENSMUST00000163610.9
ENSMUST00000164681.8 |
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr1_-_59276252 | 0.29 |
ENSMUST00000163058.2
ENSMUST00000160945.2 ENSMUST00000027178.13 |
Als2
|
alsin Rho guanine nucleotide exchange factor |
| chr10_+_126899468 | 0.29 |
ENSMUST00000120226.8
ENSMUST00000133115.8 |
Cdk4
|
cyclin-dependent kinase 4 |
| chr10_-_75673175 | 0.29 |
ENSMUST00000220440.2
|
Gstt2
|
glutathione S-transferase, theta 2 |
| chr6_-_113717689 | 0.29 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr7_-_16761732 | 0.27 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr7_+_127187910 | 0.27 |
ENSMUST00000205694.2
ENSMUST00000033088.8 ENSMUST00000206914.2 |
Rnf40
|
ring finger protein 40 |
| chr14_-_55880980 | 0.26 |
ENSMUST00000132338.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr15_+_84926909 | 0.26 |
ENSMUST00000229203.2
|
Fam118a
|
family with sequence similarity 118, member A |
| chr16_+_3726665 | 0.26 |
ENSMUST00000145150.9
ENSMUST00000040881.14 ENSMUST00000124849.2 |
Cluap1
|
clusterin associated protein 1 |
| chrX_-_7999009 | 0.26 |
ENSMUST00000130832.8
ENSMUST00000033506.13 ENSMUST00000115623.8 ENSMUST00000153839.2 |
Wdr13
|
WD repeat domain 13 |
| chr1_-_59158902 | 0.26 |
ENSMUST00000087475.11
|
Tmem237
|
transmembrane protein 237 |
| chr16_+_32238520 | 0.26 |
ENSMUST00000014220.15
ENSMUST00000080316.8 |
Dynlt2b
|
dynein light chain Tctex-type 2B |
| chr9_+_65368207 | 0.26 |
ENSMUST00000034955.8
ENSMUST00000213957.2 |
Spg21
|
SPG21, maspardin |
| chr13_-_107073415 | 0.26 |
ENSMUST00000080856.14
|
Ipo11
|
importin 11 |
| chr7_+_140461860 | 0.26 |
ENSMUST00000026560.14
|
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chrX_+_47712676 | 0.26 |
ENSMUST00000177710.2
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr1_+_75498162 | 0.26 |
ENSMUST00000027414.16
ENSMUST00000113553.2 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr14_-_30723292 | 0.25 |
ENSMUST00000228736.2
ENSMUST00000226374.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr7_-_133378410 | 0.25 |
ENSMUST00000130182.2
ENSMUST00000106139.8 |
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr3_-_105867415 | 0.25 |
ENSMUST00000118209.8
|
Atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr8_+_110944575 | 0.25 |
ENSMUST00000056972.6
|
Cmtr2
|
cap methyltransferase 2 |
| chr5_-_114961501 | 0.24 |
ENSMUST00000100850.6
|
1500011B03Rik
|
RIKEN cDNA 1500011B03 gene |
| chr10_-_79710468 | 0.24 |
ENSMUST00000092325.11
|
Plppr3
|
phospholipid phosphatase related 3 |
| chr8_-_84059048 | 0.24 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr4_-_133694543 | 0.24 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr2_+_166634445 | 0.24 |
ENSMUST00000065753.2
|
Trp53rkb
|
transformation related protein 53 regulating kinase B |
| chr16_+_49519561 | 0.24 |
ENSMUST00000046777.11
ENSMUST00000142682.9 |
Ift57
|
intraflagellar transport 57 |
| chr4_-_116982804 | 0.23 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr11_+_70021155 | 0.23 |
ENSMUST00000041550.12
ENSMUST00000165951.8 |
Mgl2
|
macrophage galactose N-acetyl-galactosamine specific lectin 2 |
| chr5_-_149559636 | 0.23 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_-_89152320 | 0.23 |
ENSMUST00000107464.8
ENSMUST00000090924.13 |
Trim46
|
tripartite motif-containing 46 |
| chr7_-_79882228 | 0.23 |
ENSMUST00000123279.8
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr12_-_110855521 | 0.23 |
ENSMUST00000220607.2
ENSMUST00000221182.2 ENSMUST00000222457.2 ENSMUST00000221102.2 ENSMUST00000043716.9 |
Cinp
|
cyclin-dependent kinase 2 interacting protein |
| chr1_+_75213044 | 0.23 |
ENSMUST00000188931.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr9_-_24414423 | 0.23 |
ENSMUST00000142064.8
ENSMUST00000170356.2 |
Dpy19l1
|
dpy-19-like 1 (C. elegans) |
| chr11_-_87249837 | 0.23 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr12_+_72583114 | 0.23 |
ENSMUST00000044352.8
|
Pcnx4
|
pecanex homolog 4 |
| chr19_-_10533562 | 0.23 |
ENSMUST00000025569.9
|
Tmem216
|
transmembrane protein 216 |
| chr17_+_36290743 | 0.23 |
ENSMUST00000087200.4
|
Gnl1
|
guanine nucleotide binding protein-like 1 |
| chr8_-_79547707 | 0.23 |
ENSMUST00000130325.8
ENSMUST00000051867.7 |
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_-_41314877 | 0.22 |
ENSMUST00000030145.9
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr5_-_149559667 | 0.22 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr7_-_109092834 | 0.22 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chr14_-_70391260 | 0.22 |
ENSMUST00000035612.7
|
Ccar2
|
cell cycle activator and apoptosis regulator 2 |
| chr9_+_20563386 | 0.22 |
ENSMUST00000034689.8
|
Pin1
|
protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1 |
| chr2_+_90735077 | 0.22 |
ENSMUST00000111464.8
ENSMUST00000090682.4 |
Kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chrX_+_47712614 | 0.22 |
ENSMUST00000114936.8
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr7_+_126446588 | 0.22 |
ENSMUST00000141805.8
ENSMUST00000064110.14 ENSMUST00000205938.2 ENSMUST00000152051.8 |
Doc2a
|
double C2, alpha |
| chr15_+_59186876 | 0.22 |
ENSMUST00000022977.14
ENSMUST00000100640.5 |
Sqle
|
squalene epoxidase |
| chr17_+_35191661 | 0.22 |
ENSMUST00000007248.5
|
Hspa1l
|
heat shock protein 1-like |
| chr2_+_164587948 | 0.22 |
ENSMUST00000109327.4
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr2_-_131001916 | 0.22 |
ENSMUST00000103188.10
ENSMUST00000133602.8 ENSMUST00000028800.12 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr19_-_41884599 | 0.22 |
ENSMUST00000038677.5
|
Rrp12
|
ribosomal RNA processing 12 homolog |
| chr11_-_59730654 | 0.21 |
ENSMUST00000019517.10
|
Cops3
|
COP9 signalosome subunit 3 |
| chr16_-_91525655 | 0.21 |
ENSMUST00000117644.8
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr6_-_83030759 | 0.21 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr5_+_138170259 | 0.21 |
ENSMUST00000019662.11
ENSMUST00000151318.8 |
Ap4m1
|
adaptor-related protein complex AP-4, mu 1 |
| chr2_+_125994050 | 0.21 |
ENSMUST00000170908.8
|
Dtwd1
|
DTW domain containing 1 |
| chr17_-_45903410 | 0.21 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr15_+_80832685 | 0.21 |
ENSMUST00000023043.10
ENSMUST00000164806.6 ENSMUST00000207170.2 ENSMUST00000168756.8 |
Adsl
|
adenylosuccinate lyase |
| chr4_-_155753628 | 0.21 |
ENSMUST00000103176.10
|
Mib2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr14_-_56339915 | 0.20 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr13_-_100912308 | 0.20 |
ENSMUST00000075550.4
|
Cenph
|
centromere protein H |
| chr8_-_84831391 | 0.20 |
ENSMUST00000041367.9
ENSMUST00000210279.2 |
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr14_-_86986541 | 0.20 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr10_+_39009951 | 0.20 |
ENSMUST00000019991.8
|
Tube1
|
tubulin, epsilon 1 |
| chr16_-_91525863 | 0.20 |
ENSMUST00000073466.13
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr12_-_84455764 | 0.20 |
ENSMUST00000120942.8
ENSMUST00000110272.9 |
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr15_-_103123711 | 0.19 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr15_-_79025387 | 0.19 |
ENSMUST00000187550.7
ENSMUST00000188562.7 ENSMUST00000190509.7 ENSMUST00000190730.7 ENSMUST00000190959.7 ENSMUST00000169604.8 ENSMUST00000186053.7 |
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chr3_+_37366662 | 0.19 |
ENSMUST00000138710.3
ENSMUST00000057975.8 ENSMUST00000108121.4 |
Gm43439
Bbs12
|
predicted gene 43439 Bardet-Biedl syndrome 12 (human) |
| chr3_-_107992662 | 0.19 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr14_-_55873026 | 0.19 |
ENSMUST00000125133.2
ENSMUST00000047131.16 |
Ipo4
|
importin 4 |
| chr8_-_23295603 | 0.19 |
ENSMUST00000163739.3
ENSMUST00000210656.2 |
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr1_+_181952302 | 0.19 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr15_+_5173342 | 0.19 |
ENSMUST00000051186.9
ENSMUST00000228218.2 |
Prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr7_+_16550198 | 0.19 |
ENSMUST00000108495.9
|
Strn4
|
striatin, calmodulin binding protein 4 |
| chr4_+_118478357 | 0.19 |
ENSMUST00000147373.2
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr7_-_80550968 | 0.19 |
ENSMUST00000146402.2
ENSMUST00000026816.15 |
Wdr73
|
WD repeat domain 73 |
| chr4_-_56802266 | 0.18 |
ENSMUST00000030140.3
|
Elp1
|
elongator complex protein 1 |
| chr5_-_138170644 | 0.18 |
ENSMUST00000000505.16
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_+_21629932 | 0.18 |
ENSMUST00000056045.5
|
Fam185a
|
family with sequence similarity 185, member A |
| chr13_-_30170031 | 0.18 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr17_+_56611313 | 0.18 |
ENSMUST00000113035.8
ENSMUST00000113039.9 ENSMUST00000142387.2 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr15_+_79113341 | 0.18 |
ENSMUST00000163571.8
|
Pick1
|
protein interacting with C kinase 1 |
| chr16_+_35861554 | 0.18 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chr15_-_85918378 | 0.18 |
ENSMUST00000016172.10
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr9_+_107926441 | 0.17 |
ENSMUST00000112295.9
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr9_+_63509925 | 0.17 |
ENSMUST00000041551.9
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr9_-_106563091 | 0.17 |
ENSMUST00000046735.11
|
Tex264
|
testis expressed gene 264 |
| chr11_+_22462088 | 0.17 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr19_+_6111204 | 0.17 |
ENSMUST00000162726.5
|
Znhit2
|
zinc finger, HIT domain containing 2 |
| chr3_+_105866685 | 0.17 |
ENSMUST00000010278.12
|
Wdr77
|
WD repeat domain 77 |
| chr1_+_90531183 | 0.17 |
ENSMUST00000186750.2
|
Cops8
|
COP9 signalosome subunit 8 |
| chr8_+_85786684 | 0.17 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr19_-_7218363 | 0.17 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr11_+_3845221 | 0.17 |
ENSMUST00000109996.8
ENSMUST00000055931.5 |
Dusp18
|
dual specificity phosphatase 18 |
| chr15_+_78312764 | 0.17 |
ENSMUST00000162517.8
ENSMUST00000166142.10 ENSMUST00000089414.11 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr2_-_35939377 | 0.17 |
ENSMUST00000070112.6
|
Ndufa8
|
NADH:ubiquinone oxidoreductase subunit A8 |
| chr17_-_56916771 | 0.17 |
ENSMUST00000052832.6
|
Micos13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr4_-_154059495 | 0.17 |
ENSMUST00000030893.3
|
Dffb
|
DNA fragmentation factor, beta subunit |
| chr17_+_79244553 | 0.16 |
ENSMUST00000024887.6
ENSMUST00000233068.2 ENSMUST00000233777.2 |
Ndufaf7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chr17_-_34243608 | 0.16 |
ENSMUST00000025183.9
|
Ring1
|
ring finger protein 1 |
| chr7_-_79882313 | 0.16 |
ENSMUST00000206084.2
ENSMUST00000205996.2 ENSMUST00000071457.12 |
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr6_+_117883732 | 0.16 |
ENSMUST00000179224.8
ENSMUST00000035493.14 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr11_+_87938128 | 0.16 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr16_-_93400691 | 0.16 |
ENSMUST00000023669.14
ENSMUST00000233931.2 ENSMUST00000154355.3 |
Setd4
|
SET domain containing 4 |
| chr10_-_68377672 | 0.16 |
ENSMUST00000020103.9
|
Cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr2_-_131016573 | 0.16 |
ENSMUST00000127987.2
|
Spef1
|
sperm flagellar 1 |
| chr18_-_24153805 | 0.16 |
ENSMUST00000066497.12
|
Zfp24
|
zinc finger protein 24 |
| chr14_+_47605208 | 0.16 |
ENSMUST00000151405.9
|
Lgals3
|
lectin, galactose binding, soluble 3 |
| chr11_+_109541747 | 0.16 |
ENSMUST00000049527.7
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr10_-_62414236 | 0.16 |
ENSMUST00000065887.14
|
Kifbp
|
kinesin family binding protein |
| chr1_-_58735106 | 0.16 |
ENSMUST00000055313.14
ENSMUST00000188772.7 |
Flacc1
|
flagellum associated containing coiled-coil domains 1 |
| chr16_-_4782031 | 0.16 |
ENSMUST00000023157.6
ENSMUST00000229765.2 |
Anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr1_-_75156993 | 0.16 |
ENSMUST00000027396.15
|
Abcb6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr15_+_102240134 | 0.16 |
ENSMUST00000113682.9
ENSMUST00000001331.13 ENSMUST00000171244.2 |
Myg1
|
melanocyte proliferating gene 1 |
| chr11_-_74787877 | 0.16 |
ENSMUST00000057631.12
ENSMUST00000081799.6 |
Sgsm2
|
small G protein signaling modulator 2 |
| chr15_+_78312851 | 0.16 |
ENSMUST00000159771.8
|
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr15_+_25774070 | 0.16 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr1_+_135945798 | 0.16 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr9_+_82711859 | 0.16 |
ENSMUST00000034783.6
|
Irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr7_-_133378468 | 0.16 |
ENSMUST00000033290.12
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr3_+_89970088 | 0.16 |
ENSMUST00000238911.2
ENSMUST00000029551.3 |
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chr11_-_101442663 | 0.16 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr7_-_98790275 | 0.15 |
ENSMUST00000037968.10
|
Uvrag
|
UV radiation resistance associated gene |
| chr4_+_130001349 | 0.15 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr5_-_129085558 | 0.15 |
ENSMUST00000100680.10
|
Stx2
|
syntaxin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx3_Rfx1_Rfx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.2 | 0.7 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.4 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.8 | GO:0035026 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.5 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.4 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.4 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.3 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.1 | 0.8 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.5 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.1 | 0.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.7 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.4 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.2 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.4 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.3 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:2000521 | positive regulation of mononuclear cell migration(GO:0071677) negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.4 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0099542 | diacylglycerol catabolic process(GO:0046340) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.3 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.4 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:2000331 | positive regulation of Wnt protein secretion(GO:0061357) regulation of terminal button organization(GO:2000331) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.2 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.0 | 0.1 | GO:0071609 | negative regulation of mitochondrial fusion(GO:0010637) negative regulation of defense response to virus by host(GO:0050689) chemokine (C-C motif) ligand 5 production(GO:0071609) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of macromitophagy(GO:1901526) regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:1904736 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.0 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.5 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.4 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0090345 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 0.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 1.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 0.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 1.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.7 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.5 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.7 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.2 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.0 | 0.1 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.1 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 1.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.4 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.0 | GO:0047726 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 1.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.1 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | Genes involved in Assembly of the pre-replicative complex |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |