Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Rfx5
Z-value: 2.61
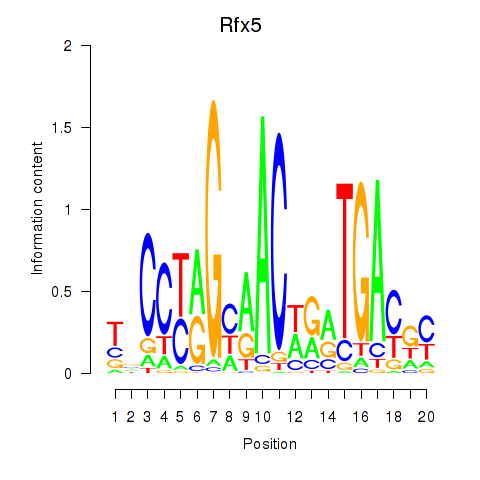
Transcription factors associated with Rfx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx5
|
ENSMUSG00000005774.13 | regulatory factor X, 5 (influences HLA class II expression) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx5 | mm39_v1_chr3_+_94861386_94861537 | 0.45 | 4.4e-01 | Click! |
Activity profile of Rfx5 motif
Sorted Z-values of Rfx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_35658131 | 4.48 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr17_+_35598583 | 2.92 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr17_-_34406193 | 2.84 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr17_+_34416707 | 2.45 |
ENSMUST00000025196.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr17_-_34219225 | 2.45 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr17_+_34416689 | 2.26 |
ENSMUST00000173441.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr7_+_46490899 | 1.74 |
ENSMUST00000147535.8
|
Ldha
|
lactate dehydrogenase A |
| chr17_+_34406762 | 1.74 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr17_+_37581103 | 1.74 |
ENSMUST00000038580.7
|
H2-M3
|
histocompatibility 2, M region locus 3 |
| chr17_+_34406523 | 1.63 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_-_123663440 | 1.60 |
ENSMUST00000197682.6
|
Gm49027
|
predicted gene, 49027 |
| chr7_-_30364394 | 1.35 |
ENSMUST00000019697.9
|
Haus5
|
HAUS augmin-like complex, subunit 5 |
| chr2_-_152239966 | 1.25 |
ENSMUST00000063332.9
ENSMUST00000182625.2 |
Sox12
|
SRY (sex determining region Y)-box 12 |
| chr11_+_106167541 | 0.97 |
ENSMUST00000044462.4
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr9_-_21202353 | 0.94 |
ENSMUST00000086374.8
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr19_+_37364791 | 0.92 |
ENSMUST00000012587.4
|
Kif11
|
kinesin family member 11 |
| chr17_-_25105277 | 0.91 |
ENSMUST00000234583.2
ENSMUST00000234968.2 ENSMUST00000044252.7 |
Nubp2
|
nucleotide binding protein 2 |
| chr10_-_7539009 | 0.91 |
ENSMUST00000163085.8
ENSMUST00000159917.8 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr8_+_27532623 | 0.91 |
ENSMUST00000209856.2
ENSMUST00000098851.12 ENSMUST00000211393.2 ENSMUST00000211518.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr8_+_94941192 | 0.89 |
ENSMUST00000079961.14
ENSMUST00000212824.2 |
Nup93
|
nucleoporin 93 |
| chr7_+_101027390 | 0.84 |
ENSMUST00000084895.12
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chrX_-_158921370 | 0.83 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr17_+_25105617 | 0.82 |
ENSMUST00000117890.8
ENSMUST00000168265.8 ENSMUST00000120943.8 ENSMUST00000068508.13 ENSMUST00000119829.8 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr7_+_100186399 | 0.79 |
ENSMUST00000120454.3
|
Coa4
|
cytochrome c oxidase assembly factor 4 |
| chr1_-_155022501 | 0.76 |
ENSMUST00000027744.10
|
Mr1
|
major histocompatibility complex, class I-related |
| chr10_-_7539333 | 0.76 |
ENSMUST00000162606.8
|
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr9_-_21202693 | 0.72 |
ENSMUST00000213407.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr6_+_94477294 | 0.71 |
ENSMUST00000061118.11
|
Slc25a26
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| chr8_+_27532583 | 0.67 |
ENSMUST00000033875.10
ENSMUST00000209525.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr17_+_34524884 | 0.63 |
ENSMUST00000074557.11
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr5_-_140346132 | 0.63 |
ENSMUST00000196130.5
|
Snx8
|
sorting nexin 8 |
| chr4_-_116485118 | 0.56 |
ENSMUST00000030456.14
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr13_+_25015653 | 0.48 |
ENSMUST00000038039.3
ENSMUST00000223804.2 |
Tdp2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr19_+_34194990 | 0.47 |
ENSMUST00000119603.2
|
Stambpl1
|
STAM binding protein like 1 |
| chr1_+_134333720 | 0.47 |
ENSMUST00000173908.8
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr2_-_92264374 | 0.46 |
ENSMUST00000111278.2
ENSMUST00000090559.12 |
Cry2
|
cryptochrome 2 (photolyase-like) |
| chr9_-_21202545 | 0.46 |
ENSMUST00000215619.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr11_+_98573377 | 0.43 |
ENSMUST00000017365.15
|
Psmd3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr5_-_125371162 | 0.43 |
ENSMUST00000127148.2
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr1_-_182929025 | 0.41 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr16_-_31898088 | 0.40 |
ENSMUST00000023467.9
|
Pak2
|
p21 (RAC1) activated kinase 2 |
| chr17_+_34482183 | 0.32 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr4_+_137989783 | 0.30 |
ENSMUST00000105821.3
|
Kif17
|
kinesin family member 17 |
| chr5_-_134485081 | 0.28 |
ENSMUST00000111244.5
|
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr17_+_34524841 | 0.26 |
ENSMUST00000235530.2
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr1_+_194302123 | 0.22 |
ENSMUST00000027952.12
|
Plxna2
|
plexin A2 |
| chr11_+_61395964 | 0.21 |
ENSMUST00000102657.10
|
B9d1
|
B9 protein domain 1 |
| chr1_+_134333506 | 0.19 |
ENSMUST00000027726.14
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr7_-_101570393 | 0.17 |
ENSMUST00000106965.8
ENSMUST00000106968.8 ENSMUST00000106967.8 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr17_+_34372046 | 0.16 |
ENSMUST00000114232.4
|
H2-DMb1
|
histocompatibility 2, class II, locus Mb1 |
| chr10_-_123032821 | 0.15 |
ENSMUST00000219619.2
ENSMUST00000020334.9 |
Usp15
|
ubiquitin specific peptidase 15 |
| chr6_-_29164981 | 0.14 |
ENSMUST00000007993.16
|
Rbm28
|
RNA binding motif protein 28 |
| chr7_-_101570382 | 0.11 |
ENSMUST00000098236.9
|
Lrrc51
|
leucine rich repeat containing 51 |
| chr19_-_4665509 | 0.11 |
ENSMUST00000053597.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr10_-_78143384 | 0.11 |
ENSMUST00000150828.2
ENSMUST00000239481.2 |
Agpat3
ENSMUSG00000118646.2
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr19_-_4665668 | 0.10 |
ENSMUST00000113822.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr4_+_66745803 | 0.09 |
ENSMUST00000048096.12
ENSMUST00000107365.3 |
Tlr4
|
toll-like receptor 4 |
| chr17_+_34457868 | 0.08 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr14_-_50322136 | 0.08 |
ENSMUST00000072370.3
|
Olfr726
|
olfactory receptor 726 |
| chr4_-_35845204 | 0.08 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr9_+_44684324 | 0.07 |
ENSMUST00000214854.3
ENSMUST00000125877.8 |
Ift46
|
intraflagellar transport 46 |
| chr1_-_133681419 | 0.07 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr7_+_100186259 | 0.06 |
ENSMUST00000054310.4
|
Coa4
|
cytochrome c oxidase assembly factor 4 |
| chr14_+_53607470 | 0.06 |
ENSMUST00000103652.5
|
Trav14n-3
|
T cell receptor alpha variable 14N-3 |
| chr10_+_78749005 | 0.05 |
ENSMUST00000204587.4
ENSMUST00000217073.3 |
Olfr1354
|
olfactory receptor 1354 |
| chr2_+_88323664 | 0.04 |
ENSMUST00000075025.7
|
Olfr1185-ps1
|
olfactory receptor 1185, pseudogene 1 |
| chr11_-_99689758 | 0.04 |
ENSMUST00000105057.2
|
Gm11569
|
predicted gene 11569 |
| chr8_+_82582953 | 0.04 |
ENSMUST00000109851.3
|
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr4_-_112291169 | 0.04 |
ENSMUST00000058605.3
|
Skint9
|
selection and upkeep of intraepithelial T cells 9 |
| chr5_+_104318542 | 0.04 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr7_+_106443820 | 0.03 |
ENSMUST00000098142.4
|
Olfr703
|
olfactory receptor 703 |
| chr6_-_124756645 | 0.03 |
ENSMUST00000147669.2
ENSMUST00000128697.8 ENSMUST00000032218.10 ENSMUST00000112475.9 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr3_+_16237371 | 0.03 |
ENSMUST00000108345.9
ENSMUST00000191774.6 ENSMUST00000108346.5 |
Ythdf3
|
YTH N6-methyladenosine RNA binding protein 3 |
| chr5_-_145816774 | 0.02 |
ENSMUST00000035918.8
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr13_-_23673000 | 0.02 |
ENSMUST00000223877.2
ENSMUST00000041541.11 ENSMUST00000110432.2 ENSMUST00000110433.10 |
Btn2a2
|
butyrophilin, subfamily 2, member A2 |
| chr10_+_90665270 | 0.02 |
ENSMUST00000182202.8
ENSMUST00000182966.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_90665399 | 0.02 |
ENSMUST00000179694.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr7_+_54485336 | 0.02 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr8_+_41276027 | 0.02 |
ENSMUST00000066814.7
|
Adam39
|
a disintegrin and metallopeptidase domain 39 |
| chr5_+_11234174 | 0.01 |
ENSMUST00000168407.3
|
Gm5861
|
predicted gene 5861 |
| chr7_-_31910570 | 0.01 |
ENSMUST00000188293.2
|
Scgb2b11
|
secretoglobin, family 2B, member 11 |
| chr14_+_111912529 | 0.01 |
ENSMUST00000042767.9
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr5_+_11178835 | 0.01 |
ENSMUST00000178863.2
|
Gm8879
|
predicted gene 8879 |
| chr5_+_11733203 | 0.01 |
ENSMUST00000178989.2
|
Gm8922
|
predicted gene 8922 |
| chr2_-_28453374 | 0.01 |
ENSMUST00000028161.6
|
Cel
|
carboxyl ester lipase |
| chr5_+_11467593 | 0.01 |
ENSMUST00000179679.2
|
Gm8897
|
predicted gene 8897 |
| chr5_+_11392646 | 0.01 |
ENSMUST00000179482.2
|
Speer1
|
spermatogenesis associated glutamate (E)-rich protein 1 |
| chr5_+_11821653 | 0.01 |
ENSMUST00000178158.2
|
Gm8926
|
predicted gene 8926 |
| chr5_+_11307124 | 0.01 |
ENSMUST00000177727.2
|
Gm8890
|
predicted gene 8890 |
| chr7_-_32873289 | 0.01 |
ENSMUST00000186529.2
|
Scgb2b18
|
secretoglobin, family 2B, member 18 |
| chr4_-_126219465 | 0.01 |
ENSMUST00000102616.8
|
Tekt2
|
tektin 2 |
| chr7_+_80764564 | 0.00 |
ENSMUST00000119083.2
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr1_+_88139678 | 0.00 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr7_-_32529087 | 0.00 |
ENSMUST00000182975.2
|
Scgb2b15
|
secretoglobin, family 2B, member 15 |
| chr1_-_53031814 | 0.00 |
ENSMUST00000190831.7
ENSMUST00000190726.2 |
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr8_+_41246310 | 0.00 |
ENSMUST00000056331.8
|
Adam20
|
a disintegrin and metallopeptidase domain 20 |
| chr14_+_53886861 | 0.00 |
ENSMUST00000103660.4
|
Trav15-2-dv6-2
|
T cell receptor alpha variable 15-2-DV6-2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.9 | 1.7 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.6 | 2.4 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.2 | 0.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 1.7 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 | 0.5 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 2.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.4 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 6.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.9 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.3 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.7 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 0.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 5.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 0.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.8 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:0071460 | positive regulation of enamel mineralization(GO:0070175) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 1.4 | GO:0051225 | spindle assembly(GO:0051225) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.6 | 9.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 3.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 2.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.4 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.7 | 9.9 | GO:0046977 | TAP binding(GO:0046977) |
| 0.3 | 1.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 7.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 0.8 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 1.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 2.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.5 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 1.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 2.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 2.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |