Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Rhox11
Z-value: 0.64
Transcription factors associated with Rhox11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rhox11
|
ENSMUSG00000051038.11 | reproductive homeobox 11 |
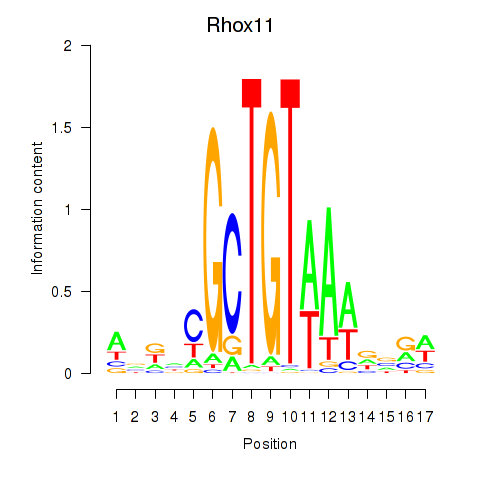
Activity profile of Rhox11 motif
Sorted Z-values of Rhox11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_138926577 | 0.74 |
ENSMUST00000145368.8
|
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr9_+_110867807 | 0.55 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr10_-_127147609 | 0.39 |
ENSMUST00000037290.12
ENSMUST00000171564.8 |
Mars1
|
methionine-tRNA synthetase 1 |
| chr11_+_23234644 | 0.37 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr1_-_169796709 | 0.34 |
ENSMUST00000027989.13
ENSMUST00000111353.4 |
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr16_+_22926162 | 0.33 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr8_+_126456710 | 0.33 |
ENSMUST00000143504.8
|
Ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr15_-_79658608 | 0.29 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr10_+_58230203 | 0.26 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_-_46558894 | 0.26 |
ENSMUST00000142706.9
ENSMUST00000173349.8 ENSMUST00000087026.13 |
Polr1c
|
polymerase (RNA) I polypeptide C |
| chr16_+_20470402 | 0.26 |
ENSMUST00000007212.9
ENSMUST00000232629.2 |
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr10_-_59787646 | 0.24 |
ENSMUST00000020308.5
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chrX_+_37689503 | 0.24 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr10_-_77845571 | 0.23 |
ENSMUST00000020522.9
|
Pfkl
|
phosphofructokinase, liver, B-type |
| chr11_-_69786324 | 0.22 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr6_+_137731599 | 0.21 |
ENSMUST00000204356.2
|
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr9_+_98178646 | 0.21 |
ENSMUST00000112938.8
ENSMUST00000112937.3 |
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr15_-_79658584 | 0.19 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr6_-_129600798 | 0.19 |
ENSMUST00000095412.10
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr9_-_78396407 | 0.19 |
ENSMUST00000154207.8
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr9_+_98178608 | 0.19 |
ENSMUST00000112935.8
|
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr11_+_98632631 | 0.18 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr6_+_137731526 | 0.18 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr19_+_57441332 | 0.17 |
ENSMUST00000026073.14
ENSMUST00000026072.5 ENSMUST00000238107.2 |
Trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr14_+_14296748 | 0.17 |
ENSMUST00000022268.10
|
Pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr16_+_22926504 | 0.16 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr7_-_30364394 | 0.16 |
ENSMUST00000019697.9
|
Haus5
|
HAUS augmin-like complex, subunit 5 |
| chr17_+_41121979 | 0.15 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr6_-_129600812 | 0.15 |
ENSMUST00000168919.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr12_-_99849660 | 0.15 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr10_+_58230183 | 0.14 |
ENSMUST00000020077.11
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr1_+_132433968 | 0.13 |
ENSMUST00000058167.3
|
Tmem81
|
transmembrane protein 81 |
| chr11_-_102771806 | 0.13 |
ENSMUST00000107060.8
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr11_-_102771751 | 0.12 |
ENSMUST00000021306.14
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr9_-_35030479 | 0.12 |
ENSMUST00000213526.2
ENSMUST00000215089.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr4_+_86493905 | 0.12 |
ENSMUST00000091064.8
|
Rraga
|
Ras-related GTP binding A |
| chr14_-_47059694 | 0.11 |
ENSMUST00000111817.8
ENSMUST00000079314.12 |
Gmfb
|
glia maturation factor, beta |
| chr9_-_107167046 | 0.11 |
ENSMUST00000035194.8
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr9_-_44624496 | 0.11 |
ENSMUST00000144251.8
ENSMUST00000156918.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr2_+_35512172 | 0.10 |
ENSMUST00000112992.9
|
Dab2ip
|
disabled 2 interacting protein |
| chrX_+_104123367 | 0.10 |
ENSMUST00000119477.2
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr13_+_34151084 | 0.09 |
ENSMUST00000222740.2
|
Nqo2
|
N-ribosyldihydronicotinamide quinone reductase 2 |
| chr18_-_56705960 | 0.09 |
ENSMUST00000174518.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr4_+_126450728 | 0.09 |
ENSMUST00000048391.15
|
Clspn
|
claspin |
| chr4_+_155048571 | 0.08 |
ENSMUST00000030931.11
ENSMUST00000070953.11 |
Pank4
|
pantothenate kinase 4 |
| chr1_-_43866910 | 0.08 |
ENSMUST00000153317.6
ENSMUST00000128261.2 ENSMUST00000126008.8 ENSMUST00000139451.8 |
Uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr18_+_63110924 | 0.08 |
ENSMUST00000150267.2
ENSMUST00000236925.2 |
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr3_-_51184730 | 0.08 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr15_-_38079089 | 0.07 |
ENSMUST00000110336.4
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr18_+_63110899 | 0.07 |
ENSMUST00000025474.14
|
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr16_+_29398149 | 0.07 |
ENSMUST00000160597.8
|
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr3_-_51184895 | 0.07 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr2_-_5867734 | 0.07 |
ENSMUST00000071016.3
|
Gm13199
|
predicted gene 13199 |
| chr18_-_42084249 | 0.07 |
ENSMUST00000070949.6
ENSMUST00000235606.2 |
Prelid2
|
PRELI domain containing 2 |
| chr6_-_106777014 | 0.06 |
ENSMUST00000013882.10
ENSMUST00000113239.10 |
Crbn
|
cereblon |
| chrX_-_101200670 | 0.06 |
ENSMUST00000056904.3
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr13_-_3661064 | 0.06 |
ENSMUST00000096069.5
|
Tasor2
|
transcription activation suppressor family member 2 |
| chrX_+_104123341 | 0.06 |
ENSMUST00000033577.11
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr8_+_22996233 | 0.06 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr5_+_21391282 | 0.06 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr8_+_14938022 | 0.06 |
ENSMUST00000123990.2
ENSMUST00000027554.8 |
Cln8
|
CLN8 transmembrane ER and ERGIC protein |
| chr17_+_46558995 | 0.06 |
ENSMUST00000095263.10
ENSMUST00000123311.8 |
Yipf3
|
Yip1 domain family, member 3 |
| chr12_-_114398864 | 0.05 |
ENSMUST00000103489.2
|
Ighv6-6
|
immunoglobulin heavy variable 6-6 |
| chr19_-_8796288 | 0.04 |
ENSMUST00000153281.2
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr1_+_153628598 | 0.04 |
ENSMUST00000182538.3
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr17_-_34736326 | 0.04 |
ENSMUST00000075483.5
|
Btnl6
|
butyrophilin-like 6 |
| chr3_+_127584449 | 0.04 |
ENSMUST00000171621.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr9_-_20432562 | 0.04 |
ENSMUST00000215908.2
ENSMUST00000068296.8 ENSMUST00000174462.8 ENSMUST00000213418.2 |
Zfp266
|
zinc finger protein 266 |
| chr12_+_76884182 | 0.04 |
ENSMUST00000041008.10
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr9_-_20556031 | 0.04 |
ENSMUST00000148631.8
ENSMUST00000131128.2 ENSMUST00000151861.9 ENSMUST00000131343.8 ENSMUST00000086458.10 |
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr8_+_89373943 | 0.03 |
ENSMUST00000118370.8
ENSMUST00000054324.15 |
Nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr7_-_46445085 | 0.03 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr3_+_68375495 | 0.03 |
ENSMUST00000182532.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr10_+_41179966 | 0.03 |
ENSMUST00000173494.4
|
Ak9
|
adenylate kinase 9 |
| chr6_-_57827328 | 0.03 |
ENSMUST00000203310.3
ENSMUST00000203488.3 |
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr6_-_129637519 | 0.03 |
ENSMUST00000119533.2
ENSMUST00000145984.8 ENSMUST00000118401.8 ENSMUST00000112057.9 ENSMUST00000071920.11 |
Klrc2
|
killer cell lectin-like receptor subfamily C, member 2 |
| chr5_+_93241287 | 0.02 |
ENSMUST00000074733.11
ENSMUST00000201700.4 ENSMUST00000202196.4 ENSMUST00000202308.4 |
Septin11
|
septin 11 |
| chr5_+_122239030 | 0.02 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr7_+_126184141 | 0.02 |
ENSMUST00000137646.8
|
Apobr
|
apolipoprotein B receptor |
| chr8_-_118398264 | 0.02 |
ENSMUST00000037955.14
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr14_-_20546848 | 0.02 |
ENSMUST00000022353.5
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr3_-_49711706 | 0.02 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr1_+_180769890 | 0.02 |
ENSMUST00000161847.8
|
Tmem63a
|
transmembrane protein 63a |
| chr3_-_49711765 | 0.02 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr16_+_29398165 | 0.01 |
ENSMUST00000161186.8
ENSMUST00000038867.13 |
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr7_+_19927635 | 0.01 |
ENSMUST00000168984.2
|
Vmn1r95
|
vomeronasal 1 receptor, 95 |
| chrX_+_151909893 | 0.01 |
ENSMUST00000163801.2
|
Foxr2
|
forkhead box R2 |
| chr6_-_123627684 | 0.01 |
ENSMUST00000170808.3
|
Vmn2r22
|
vomeronasal 2, receptor 22 |
| chr14_-_21898992 | 0.01 |
ENSMUST00000124549.9
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr7_-_44773750 | 0.01 |
ENSMUST00000211725.2
ENSMUST00000003521.10 |
Rps11
|
ribosomal protein S11 |
| chr15_+_91722524 | 0.01 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr15_+_91722458 | 0.01 |
ENSMUST00000109277.8
|
Smgc
|
submandibular gland protein C |
| chr7_+_107501834 | 0.01 |
ENSMUST00000210420.2
|
Olfr472
|
olfactory receptor 472 |
| chr3_+_29568055 | 0.01 |
ENSMUST00000140288.2
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr7_-_119078472 | 0.01 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr3_+_61269059 | 0.01 |
ENSMUST00000049064.4
|
Rap2b
|
RAP2B, member of RAS oncogene family |
| chr2_-_86754889 | 0.01 |
ENSMUST00000111574.3
|
Olfr1098
|
olfactory receptor 1098 |
| chr16_-_58860130 | 0.01 |
ENSMUST00000207673.4
|
Olfr187
|
olfactory receptor 187 |
| chr19_-_39451509 | 0.01 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr8_-_81741495 | 0.01 |
ENSMUST00000042724.8
|
Usp38
|
ubiquitin specific peptidase 38 |
| chr9_+_7502341 | 0.01 |
ENSMUST00000034488.4
|
Mmp10
|
matrix metallopeptidase 10 |
| chr9_-_16289527 | 0.00 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr10_+_102376109 | 0.00 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr7_-_104542833 | 0.00 |
ENSMUST00000081116.2
|
Olfr666
|
olfactory receptor 666 |
| chr7_-_107631548 | 0.00 |
ENSMUST00000049719.4
|
Olfr478
|
olfactory receptor 478 |
| chr17_-_37935524 | 0.00 |
ENSMUST00000072265.3
|
Olfr116
|
olfactory receptor 116 |
| chr7_+_107964542 | 0.00 |
ENSMUST00000209743.2
|
Olfr494
|
olfactory receptor 494 |
| chr4_+_62398262 | 0.00 |
ENSMUST00000030088.12
ENSMUST00000107449.4 |
Bspry
|
B-box and SPRY domain containing |
| chr6_+_78382131 | 0.00 |
ENSMUST00000023906.4
|
Reg2
|
regenerating islet-derived 2 |
| chr5_-_82271183 | 0.00 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr7_-_108493612 | 0.00 |
ENSMUST00000202706.2
ENSMUST00000084752.2 |
Olfr519
|
olfactory receptor 519 |
| chr2_+_86655007 | 0.00 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr4_-_119241024 | 0.00 |
ENSMUST00000127149.8
ENSMUST00000152879.9 ENSMUST00000238673.2 ENSMUST00000238485.2 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr16_-_19425453 | 0.00 |
ENSMUST00000078603.3
ENSMUST00000218837.2 |
Olfr170
|
olfactory receptor 170 |
| chr7_-_141794815 | 0.00 |
ENSMUST00000211591.2
|
Gm40460
|
predicted gene, 40460 |
| chr11_+_73973733 | 0.00 |
ENSMUST00000178159.2
|
Zfp616
|
zinc finger protein 616 |
| chr11_-_99996452 | 0.00 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr7_+_103246525 | 0.00 |
ENSMUST00000098197.2
|
Olfr618
|
olfactory receptor 618 |
| chr3_-_144680801 | 0.00 |
ENSMUST00000029923.10
ENSMUST00000238960.2 |
Clca4a
|
chloride channel accessory 4A |
| chr11_-_100246209 | 0.00 |
ENSMUST00000146878.3
|
Hap1
|
huntingtin-associated protein 1 |
| chr17_+_38482279 | 0.00 |
ENSMUST00000215900.2
|
Olfr134
|
olfactory receptor 134 |
| chr1_+_40844739 | 0.00 |
ENSMUST00000114765.4
|
Tmem182
|
transmembrane protein 182 |
| chr7_-_119801327 | 0.00 |
ENSMUST00000033198.6
|
Crym
|
crystallin, mu |
| chr8_-_106306477 | 0.00 |
ENSMUST00000194654.2
|
Agrp
|
agouti related neuropeptide |
| chrX_+_36640238 | 0.00 |
ENSMUST00000115188.9
|
Rhox3c
|
reproductive homeobox 3C |
| chr6_+_139564196 | 0.00 |
ENSMUST00000188066.2
ENSMUST00000190962.7 |
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr17_+_79922329 | 0.00 |
ENSMUST00000040368.3
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr19_-_12069012 | 0.00 |
ENSMUST00000220005.2
|
Olfr1426
|
olfactory receptor 1426 |
| chr7_-_142253247 | 0.00 |
ENSMUST00000105934.8
|
Ins2
|
insulin II |
| chr3_-_122828592 | 0.00 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr3_+_130411294 | 0.00 |
ENSMUST00000163620.8
|
Etnppl
|
ethanolamine phosphate phospholyase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rhox11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.3 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.1 | 0.4 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.4 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.7 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.5 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.2 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:1902523 | detection of peptidoglycan(GO:0032499) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0031597 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.1 | 0.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |