Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
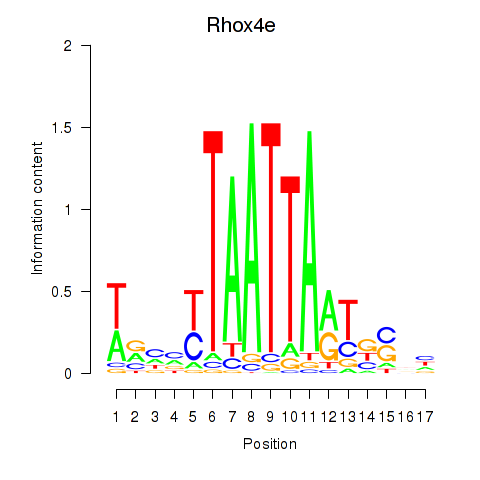
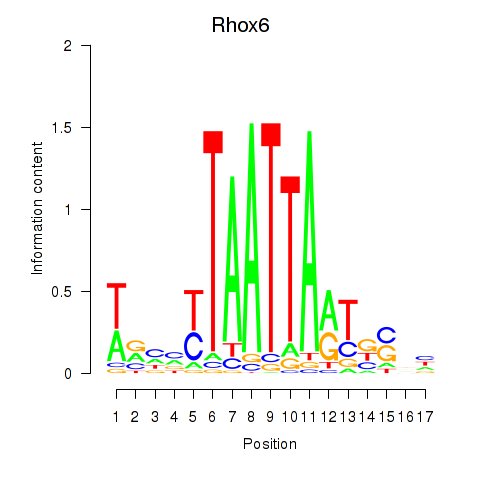
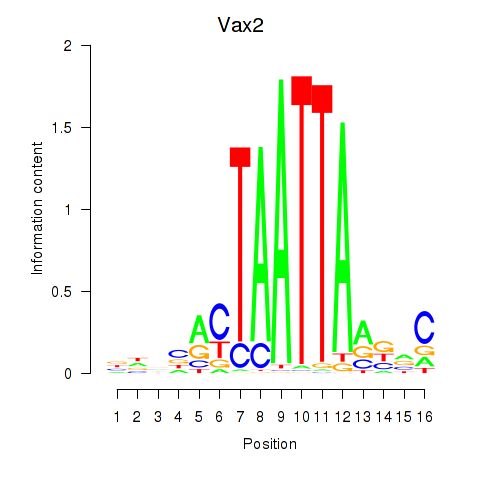
Results for Rhox4e_Rhox6_Vax2
Z-value: 1.08
Transcription factors associated with Rhox4e_Rhox6_Vax2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rhox4e
|
ENSMUSG00000071770.6 | reproductive homeobox 4E |
|
Rhox6
|
ENSMUSG00000006200.4 | reproductive homeobox 6 |
|
Vax2
|
ENSMUSG00000034777.3 | ventral anterior homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vax2 | mm39_v1_chr6_+_83688213_83688270 | 0.91 | 3.4e-02 | Click! |
| Rhox4e | mm39_v1_chrX_+_36739065_36739065 | 0.69 | 2.0e-01 | Click! |
Activity profile of Rhox4e_Rhox6_Vax2 motif
Sorted Z-values of Rhox4e_Rhox6_Vax2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_84565218 | 2.16 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr2_-_89855921 | 2.14 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr7_+_130375799 | 1.29 |
ENSMUST00000048453.7
ENSMUST00000208593.2 |
Btbd16
|
BTB (POZ) domain containing 16 |
| chr15_-_66985760 | 1.09 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr2_-_111843053 | 0.97 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chrX_+_16485937 | 0.94 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr2_-_89423470 | 0.94 |
ENSMUST00000217254.2
ENSMUST00000217192.2 ENSMUST00000213221.2 |
Olfr1246
|
olfactory receptor 1246 |
| chrX_+_149330371 | 0.91 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr13_+_23191826 | 0.85 |
ENSMUST00000228758.2
ENSMUST00000228031.2 ENSMUST00000227573.2 |
Vmn1r213
|
vomeronasal 1 receptor 213 |
| chr2_-_111820618 | 0.84 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr19_+_12364643 | 0.71 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chr3_-_129834788 | 0.70 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chrX_+_102400061 | 0.68 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr9_+_123195986 | 0.66 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr4_+_100336003 | 0.60 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr7_-_12829100 | 0.59 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr2_+_87610895 | 0.55 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152 |
| chr6_-_118456198 | 0.55 |
ENSMUST00000161170.2
|
Zfp9
|
zinc finger protein 9 |
| chr9_-_19275301 | 0.54 |
ENSMUST00000214810.2
|
Olfr846
|
olfactory receptor 846 |
| chr7_-_11414074 | 0.53 |
ENSMUST00000227010.2
ENSMUST00000209638.3 |
Vmn1r72
|
vomeronasal 1 receptor 72 |
| chr7_+_28869770 | 0.47 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr10_-_128875765 | 0.43 |
ENSMUST00000204763.3
|
Olfr764-ps1
|
olfactory receptor 764, pseudogene 1 |
| chr6_-_3399451 | 0.40 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr19_-_12313274 | 0.40 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr17_+_38456172 | 0.39 |
ENSMUST00000215078.3
ENSMUST00000215549.3 ENSMUST00000173610.2 |
Olfr133
|
olfactory receptor 133 |
| chr2_-_88994435 | 0.39 |
ENSMUST00000099793.3
|
Olfr1224-ps1
|
olfactory receptor 1224, pseudogene 1 |
| chr10_+_129539079 | 0.37 |
ENSMUST00000213331.2
|
Olfr804
|
olfactory receptor 804 |
| chr7_-_5152669 | 0.34 |
ENSMUST00000236378.2
|
Vmn1r55
|
vomeronasal 1 receptor 55 |
| chr10_-_33827185 | 0.34 |
ENSMUST00000218880.2
ENSMUST00000048222.6 ENSMUST00000218222.2 ENSMUST00000218892.2 ENSMUST00000218055.2 |
Zup1
|
zinc finger containing ubiquitin peptidase 1 |
| chr8_+_23901506 | 0.32 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr16_+_45044678 | 0.32 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr7_-_126275529 | 0.31 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr5_+_104350475 | 0.31 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr13_+_23718038 | 0.30 |
ENSMUST00000073261.3
|
H2ac10
|
H2A clustered histone 10 |
| chr7_-_106531426 | 0.29 |
ENSMUST00000215468.2
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr10_+_127226180 | 0.29 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chr3_-_15902583 | 0.29 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr14_+_15018856 | 0.28 |
ENSMUST00000169555.2
|
Gm3755
|
predicted gene 3755 |
| chr7_-_12002196 | 0.28 |
ENSMUST00000227973.2
ENSMUST00000228764.2 ENSMUST00000228482.2 ENSMUST00000227080.2 |
Vmn1r81
|
vomeronasal 1 receptor 81 |
| chr7_-_28855020 | 0.28 |
ENSMUST00000123416.2
|
Fam98c
|
family with sequence similarity 98, member C |
| chr16_-_58620631 | 0.28 |
ENSMUST00000206205.3
|
Olfr173
|
olfactory receptor 173 |
| chr9_-_39618413 | 0.28 |
ENSMUST00000215192.2
|
Olfr149
|
olfactory receptor 149 |
| chr9_+_20193647 | 0.27 |
ENSMUST00000071725.4
ENSMUST00000212983.3 |
Olfr39
|
olfactory receptor 39 |
| chr2_+_88217406 | 0.27 |
ENSMUST00000214040.3
|
Olfr1178
|
olfactory receptor 1178 |
| chr7_+_28869629 | 0.27 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr2_+_69727599 | 0.26 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr7_-_10292412 | 0.26 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr17_-_59320257 | 0.26 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr10_-_83484467 | 0.25 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr14_-_50586329 | 0.25 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr6_-_123395075 | 0.25 |
ENSMUST00000172199.3
|
Vmn2r20
|
vomeronasal 2, receptor 20 |
| chr13_-_53627110 | 0.24 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr6_-_90055488 | 0.24 |
ENSMUST00000203791.3
ENSMUST00000226368.2 |
Vmn1r49
|
vomeronasal 1, receptor 49 |
| chr6_+_90078412 | 0.23 |
ENSMUST00000089417.8
ENSMUST00000226577.2 |
Vmn1r50
|
vomeronasal 1 receptor 50 |
| chr19_+_3373285 | 0.22 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chrX_-_149372840 | 0.22 |
ENSMUST00000112725.8
ENSMUST00000112720.8 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr13_+_22508759 | 0.22 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chrX_+_159551009 | 0.21 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr2_-_111104451 | 0.20 |
ENSMUST00000214760.2
|
Olfr1277
|
olfactory receptor 1277 |
| chr17_-_37523969 | 0.20 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr10_-_44024843 | 0.20 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr13_+_23398297 | 0.19 |
ENSMUST00000236177.2
|
Vmn1r221
|
vomeronasal 1 receptor 221 |
| chr7_-_41708447 | 0.19 |
ENSMUST00000168489.3
ENSMUST00000233456.2 |
Vmn2r59
|
vomeronasal 2, receptor 59 |
| chr11_+_49410475 | 0.19 |
ENSMUST00000204706.3
|
Olfr1383
|
olfactory receptor 1383 |
| chr7_-_102507962 | 0.19 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr1_+_173093568 | 0.19 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr7_+_3648264 | 0.19 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr2_-_88157559 | 0.18 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr7_-_24705320 | 0.18 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr5_+_136023649 | 0.18 |
ENSMUST00000111142.9
ENSMUST00000111145.10 ENSMUST00000111144.8 ENSMUST00000199239.5 ENSMUST00000005072.10 ENSMUST00000130345.2 |
Dtx2
|
deltex 2, E3 ubiquitin ligase |
| chr7_+_23330147 | 0.18 |
ENSMUST00000227774.2
ENSMUST00000226771.2 ENSMUST00000228681.2 ENSMUST00000228559.2 ENSMUST00000228674.2 ENSMUST00000227866.2 ENSMUST00000227386.2 ENSMUST00000228484.2 ENSMUST00000226321.2 ENSMUST00000226128.2 ENSMUST00000226733.2 ENSMUST00000228228.2 |
Vmn1r171
|
vomeronasal 1 receptor 171 |
| chr4_+_137720326 | 0.18 |
ENSMUST00000139759.8
ENSMUST00000058133.10 ENSMUST00000105830.9 ENSMUST00000084215.12 |
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr2_-_111880531 | 0.18 |
ENSMUST00000213582.2
ENSMUST00000213961.3 ENSMUST00000215531.2 |
Olfr1312
|
olfactory receptor 1312 |
| chr8_+_45960804 | 0.17 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_-_9529898 | 0.17 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr13_-_23302027 | 0.17 |
ENSMUST00000228656.2
|
Vmn1r217
|
vomeronasal 1 receptor 217 |
| chr4_-_131802606 | 0.17 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr13_-_23302396 | 0.16 |
ENSMUST00000227110.2
|
Vmn1r217
|
vomeronasal 1 receptor 217 |
| chr2_+_3425159 | 0.16 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr10_-_128918779 | 0.15 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr7_+_17979272 | 0.15 |
ENSMUST00000066780.5
|
Mill1
|
MHC I like leukocyte 1 |
| chr12_-_114710326 | 0.15 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr6_-_41752111 | 0.15 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr7_+_86298269 | 0.14 |
ENSMUST00000214401.3
ENSMUST00000215280.2 |
Olfr293
|
olfactory receptor 293 |
| chr17_+_41121979 | 0.14 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chrM_-_14061 | 0.14 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr18_+_4993795 | 0.14 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr15_+_98350469 | 0.14 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr5_-_31684036 | 0.14 |
ENSMUST00000202421.2
ENSMUST00000201769.4 ENSMUST00000065388.11 |
Supt7l
|
SPT7-like, STAGA complex gamma subunit |
| chrM_+_14138 | 0.14 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr10_-_75946790 | 0.13 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr4_-_43710231 | 0.13 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chrX_-_142716200 | 0.13 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr2_+_172994841 | 0.12 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr9_+_40092216 | 0.12 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr4_+_134658209 | 0.12 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr11_-_99134885 | 0.12 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr18_+_4920513 | 0.12 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chrM_+_2743 | 0.12 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr12_-_114878652 | 0.12 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr7_+_12661337 | 0.11 |
ENSMUST00000045870.5
|
Rnf225
|
ring finger protein 225 |
| chr3_-_105839980 | 0.11 |
ENSMUST00000098758.5
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr10_-_81243475 | 0.11 |
ENSMUST00000140916.8
|
Nfic
|
nuclear factor I/C |
| chr12_+_80691275 | 0.11 |
ENSMUST00000217889.2
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr4_-_131802561 | 0.10 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr5_-_116162415 | 0.10 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr14_-_20546848 | 0.10 |
ENSMUST00000022353.5
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr2_+_87696836 | 0.10 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr6_+_37847721 | 0.09 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr1_+_173924457 | 0.09 |
ENSMUST00000213832.2
|
Olfr427
|
olfactory receptor 427 |
| chr8_+_114362181 | 0.09 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr5_-_137015683 | 0.09 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr2_+_85804239 | 0.09 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr2_+_87078357 | 0.09 |
ENSMUST00000214119.2
ENSMUST00000217196.3 ENSMUST00000213513.2 |
Olfr1115
|
olfactory receptor 1115 |
| chr13_+_22268610 | 0.09 |
ENSMUST00000228243.2
ENSMUST00000226680.2 |
Vmn1r188
|
vomeronasal 1 receptor 188 |
| chr9_+_38516398 | 0.09 |
ENSMUST00000217057.2
|
Olfr914
|
olfactory receptor 914 |
| chr2_+_144112798 | 0.09 |
ENSMUST00000028910.9
ENSMUST00000110027.2 |
Mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr13_+_118851214 | 0.09 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr2_+_111355548 | 0.08 |
ENSMUST00000217845.3
|
Olfr1293-ps
|
olfactory receptor 1293, pseudogene |
| chr12_+_116239006 | 0.08 |
ENSMUST00000090195.5
|
Gm11027
|
predicted gene 11027 |
| chr8_+_114362419 | 0.08 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr4_+_122910382 | 0.08 |
ENSMUST00000102649.4
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr9_-_45739886 | 0.08 |
ENSMUST00000214868.2
ENSMUST00000117194.8 |
Cep164
|
centrosomal protein 164 |
| chr14_+_54669054 | 0.08 |
ENSMUST00000089688.6
ENSMUST00000225641.2 |
Mmp14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chrX_-_74460137 | 0.07 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr7_+_126549692 | 0.07 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr2_-_125624754 | 0.07 |
ENSMUST00000053699.13
|
Secisbp2l
|
SECIS binding protein 2-like |
| chr8_+_66838927 | 0.07 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr17_-_45970238 | 0.07 |
ENSMUST00000120717.8
|
Capn11
|
calpain 11 |
| chr2_+_118877610 | 0.07 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr6_+_125529911 | 0.07 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr9_+_38382403 | 0.07 |
ENSMUST00000214377.2
|
Olfr905
|
olfactory receptor 905 |
| chr5_-_146521629 | 0.07 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chrX_+_106299484 | 0.06 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr6_+_122490534 | 0.06 |
ENSMUST00000032210.14
ENSMUST00000148517.8 |
Mfap5
|
microfibrillar associated protein 5 |
| chr16_+_56024676 | 0.06 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr2_-_89678487 | 0.06 |
ENSMUST00000214428.3
|
Olfr48
|
olfactory receptor 48 |
| chr6_-_125357756 | 0.06 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr2_-_86317730 | 0.06 |
ENSMUST00000217292.3
|
Olfr228
|
olfactory receptor 228 |
| chr9_-_19163273 | 0.05 |
ENSMUST00000214019.2
ENSMUST00000214267.2 |
Olfr843
|
olfactory receptor 843 |
| chr4_-_88595161 | 0.05 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16 |
| chr5_-_66211842 | 0.05 |
ENSMUST00000200852.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr12_+_52746158 | 0.05 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr5_-_3691453 | 0.05 |
ENSMUST00000140871.2
|
Gatad1
|
GATA zinc finger domain containing 1 |
| chr2_-_86722536 | 0.05 |
ENSMUST00000111576.3
ENSMUST00000217403.2 |
Olfr1097
|
olfactory receptor 1097 |
| chr17_-_24570003 | 0.05 |
ENSMUST00000121226.8
ENSMUST00000234232.2 |
Abca17
|
ATP-binding cassette, sub-family A (ABC1), member 17 |
| chr5_+_137015873 | 0.05 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr12_-_111150925 | 0.05 |
ENSMUST00000121608.2
|
4930595D18Rik
|
RIKEN cDNA 4930595D18 gene |
| chr5_-_138169476 | 0.05 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr1_-_131441962 | 0.04 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr4_+_19605451 | 0.04 |
ENSMUST00000108250.3
|
Gm12353
|
predicted gene 12353 |
| chr5_-_143279378 | 0.04 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr4_+_108576846 | 0.04 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr9_+_72866067 | 0.04 |
ENSMUST00000098567.9
ENSMUST00000034734.9 |
Dnaaf4
|
dynein axonemal assembly factor 4 |
| chr11_-_99447672 | 0.04 |
ENSMUST00000092699.3
|
Krtap3-2
|
keratin associated protein 3-2 |
| chr11_-_113600838 | 0.04 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr8_+_65399831 | 0.04 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr5_-_148865429 | 0.04 |
ENSMUST00000149169.3
ENSMUST00000047257.15 |
Katnal1
|
katanin p60 subunit A-like 1 |
| chr17_-_57338468 | 0.04 |
ENSMUST00000007814.10
ENSMUST00000233480.2 |
Khsrp
|
KH-type splicing regulatory protein |
| chrX_-_49517375 | 0.04 |
ENSMUST00000215270.2
|
Olfr1324
|
olfactory receptor 1324 |
| chr5_-_90788323 | 0.03 |
ENSMUST00000202784.4
ENSMUST00000031317.10 ENSMUST00000201370.2 |
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr12_-_103660916 | 0.03 |
ENSMUST00000117053.8
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr13_+_92562404 | 0.03 |
ENSMUST00000061594.13
|
Ankrd34b
|
ankyrin repeat domain 34B |
| chr4_-_133066594 | 0.03 |
ENSMUST00000043305.14
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr5_-_118382926 | 0.03 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr12_-_114443071 | 0.03 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr2_+_121787131 | 0.03 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr7_+_19833605 | 0.03 |
ENSMUST00000165330.3
|
Vmn1r91
|
vomeronasal 1 receptor 91 |
| chr4_+_137720755 | 0.03 |
ENSMUST00000084214.12
ENSMUST00000105831.9 ENSMUST00000203828.3 |
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr1_+_63215976 | 0.03 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr15_+_102927366 | 0.03 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr15_+_82140224 | 0.03 |
ENSMUST00000143238.2
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr1_-_173161069 | 0.02 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr7_+_19102423 | 0.02 |
ENSMUST00000132655.2
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr5_-_18093739 | 0.02 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr7_+_6477087 | 0.02 |
ENSMUST00000056144.7
|
Olfr1346
|
olfactory receptor 1346 |
| chr2_+_155593030 | 0.02 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr12_-_114793177 | 0.02 |
ENSMUST00000103511.2
ENSMUST00000195735.2 |
Ighv1-31
|
immunoglobulin heavy variable 1-31 |
| chr1_+_63216281 | 0.02 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chrX_+_100473161 | 0.02 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr9_+_40712562 | 0.02 |
ENSMUST00000117557.8
|
Hspa8
|
heat shock protein 8 |
| chr12_-_113649535 | 0.01 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr6_-_54949587 | 0.01 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr13_-_23041731 | 0.01 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr10_-_89270554 | 0.01 |
ENSMUST00000220071.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr5_+_13448833 | 0.01 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_87111033 | 0.01 |
ENSMUST00000044533.9
|
Prss56
|
protease, serine 56 |
| chr11_+_116734104 | 0.01 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr10_-_89270527 | 0.01 |
ENSMUST00000218764.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr5_+_24679154 | 0.01 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr6_-_68907718 | 0.01 |
ENSMUST00000114212.4
|
Igkv13-85
|
immunoglobulin kappa chain variable 13-85 |
| chr13_+_22574543 | 0.01 |
ENSMUST00000226157.2
ENSMUST00000227326.2 ENSMUST00000228726.2 |
Vmn1r200
|
vomeronasal 1 receptor 200 |
| chr16_-_88303859 | 0.01 |
ENSMUST00000069549.3
|
Cldn17
|
claudin 17 |
| chr9_+_108216233 | 0.01 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr5_-_72325482 | 0.01 |
ENSMUST00000196241.2
ENSMUST00000013693.11 |
Commd8
|
COMM domain containing 8 |
| chr1_-_192880260 | 0.01 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr1_-_154692678 | 0.01 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr18_+_34675366 | 0.01 |
ENSMUST00000012426.3
|
Wnt8a
|
wingless-type MMTV integration site family, member 8A |
| chr15_-_82678490 | 0.01 |
ENSMUST00000006094.6
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr11_+_73489420 | 0.01 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rhox4e_Rhox6_Vax2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.2 | 0.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 2.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.9 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.3 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 0.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.3 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.9 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.3 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0061033 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 1.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 5.7 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 2.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 2.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 0.7 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.2 | 2.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.2 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 2.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |