Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
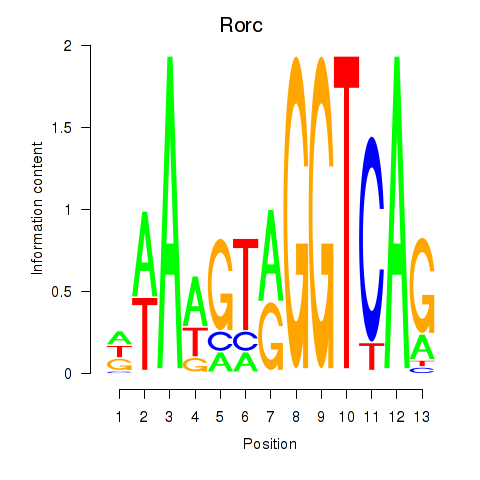
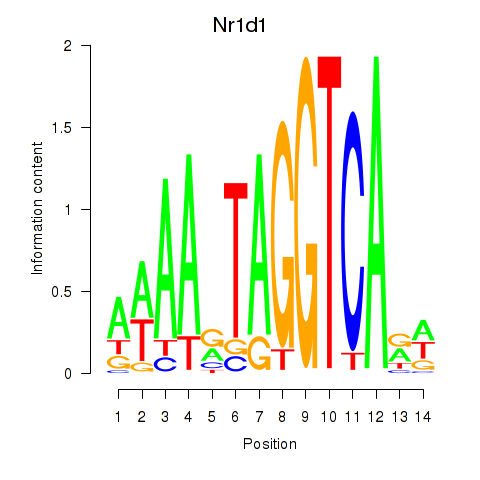
Results for Rorc_Nr1d1
Z-value: 0.44
Transcription factors associated with Rorc_Nr1d1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rorc
|
ENSMUSG00000028150.15 | RAR-related orphan receptor gamma |
|
Nr1d1
|
ENSMUSG00000020889.12 | nuclear receptor subfamily 1, group D, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1d1 | mm39_v1_chr11_-_98666159_98666183 | 0.49 | 4.1e-01 | Click! |
| Rorc | mm39_v1_chr3_+_94284812_94284834 | -0.19 | 7.5e-01 | Click! |
Activity profile of Rorc_Nr1d1 motif
Sorted Z-values of Rorc_Nr1d1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_120432669 | 0.23 |
ENSMUST00000027639.8
|
Marco
|
macrophage receptor with collagenous structure |
| chr3_-_10273628 | 0.23 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr17_+_34124078 | 0.20 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr16_-_18904240 | 0.18 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr3_-_89671888 | 0.15 |
ENSMUST00000200558.5
ENSMUST00000029562.5 |
Chrnb2
|
cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal) |
| chr17_-_46343291 | 0.14 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr17_-_46342739 | 0.14 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr2_-_48839218 | 0.11 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr17_+_25105617 | 0.11 |
ENSMUST00000117890.8
ENSMUST00000168265.8 ENSMUST00000120943.8 ENSMUST00000068508.13 ENSMUST00000119829.8 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr16_+_21828223 | 0.11 |
ENSMUST00000023561.8
|
Senp2
|
SUMO/sentrin specific peptidase 2 |
| chr2_-_86180622 | 0.10 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr1_+_151220222 | 0.10 |
ENSMUST00000023918.13
ENSMUST00000111887.10 ENSMUST00000097543.8 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr11_-_115518774 | 0.10 |
ENSMUST00000154623.2
ENSMUST00000106503.10 ENSMUST00000141614.3 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr4_+_11558905 | 0.10 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr7_-_79115915 | 0.10 |
ENSMUST00000073889.14
|
Polg
|
polymerase (DNA directed), gamma |
| chr2_+_29014106 | 0.09 |
ENSMUST00000129544.8
|
Setx
|
senataxin |
| chr12_-_34578842 | 0.09 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr3_+_108291145 | 0.09 |
ENSMUST00000090561.10
ENSMUST00000102629.8 ENSMUST00000128089.2 |
Psrc1
|
proline/serine-rich coiled-coil 1 |
| chr12_-_40298072 | 0.09 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr4_+_11579648 | 0.09 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr17_-_25105277 | 0.08 |
ENSMUST00000234583.2
ENSMUST00000234968.2 ENSMUST00000044252.7 |
Nubp2
|
nucleotide binding protein 2 |
| chr17_+_43671314 | 0.08 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr4_+_115641996 | 0.08 |
ENSMUST00000177280.8
ENSMUST00000176047.8 |
Atpaf1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr11_+_114780867 | 0.07 |
ENSMUST00000106582.9
ENSMUST00000045151.6 |
Cd300a
|
CD300A molecule |
| chr2_+_91541245 | 0.07 |
ENSMUST00000142692.2
ENSMUST00000090608.6 |
Harbi1
|
harbinger transposase derived 1 |
| chr14_+_79086492 | 0.07 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr6_-_124689001 | 0.07 |
ENSMUST00000203238.2
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr11_-_51526697 | 0.06 |
ENSMUST00000001081.10
|
Rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr5_+_145020910 | 0.06 |
ENSMUST00000124379.3
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr7_-_121620366 | 0.06 |
ENSMUST00000033160.15
|
Gga2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr2_-_152857239 | 0.06 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr12_+_3941728 | 0.05 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr3_-_53370621 | 0.05 |
ENSMUST00000056749.14
|
Nhlrc3
|
NHL repeat containing 3 |
| chr7_-_79115760 | 0.05 |
ENSMUST00000125562.2
|
Polg
|
polymerase (DNA directed), gamma |
| chr17_-_56440817 | 0.05 |
ENSMUST00000167545.3
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr8_-_85807281 | 0.05 |
ENSMUST00000152785.8
|
Wdr83
|
WD repeat domain containing 83 |
| chr8_-_85807308 | 0.05 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr11_+_96822213 | 0.05 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr2_+_153133326 | 0.05 |
ENSMUST00000028977.7
|
Kif3b
|
kinesin family member 3B |
| chr10_+_25317309 | 0.05 |
ENSMUST00000217929.2
ENSMUST00000220121.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr12_-_79237722 | 0.05 |
ENSMUST00000085254.7
|
Rdh11
|
retinol dehydrogenase 11 |
| chr2_+_96148418 | 0.05 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr2_-_48839276 | 0.05 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr19_+_6434416 | 0.05 |
ENSMUST00000035269.15
ENSMUST00000113483.2 |
Pygm
|
muscle glycogen phosphorylase |
| chr2_+_30331839 | 0.04 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr13_-_53135064 | 0.04 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr5_+_135216090 | 0.04 |
ENSMUST00000002825.6
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr5_-_136275407 | 0.04 |
ENSMUST00000196245.2
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr9_+_54771064 | 0.04 |
ENSMUST00000034843.9
|
Ireb2
|
iron responsive element binding protein 2 |
| chr15_-_81756076 | 0.04 |
ENSMUST00000023117.10
|
Phf5a
|
PHD finger protein 5A |
| chr6_-_124519240 | 0.04 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr17_+_38110779 | 0.04 |
ENSMUST00000215168.2
ENSMUST00000216478.2 |
Olfr124
|
olfactory receptor 124 |
| chr11_-_110058899 | 0.04 |
ENSMUST00000044850.4
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr11_-_115518951 | 0.04 |
ENSMUST00000155709.2
ENSMUST00000021089.11 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr13_+_52750883 | 0.04 |
ENSMUST00000055087.7
|
Syk
|
spleen tyrosine kinase |
| chr6_-_29380467 | 0.04 |
ENSMUST00000080428.13
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr7_+_13132040 | 0.04 |
ENSMUST00000005791.14
|
Cabp5
|
calcium binding protein 5 |
| chr11_+_4833186 | 0.03 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chr13_+_104365432 | 0.03 |
ENSMUST00000070761.10
ENSMUST00000225557.2 |
Cenpk
|
centromere protein K |
| chr15_+_10486092 | 0.03 |
ENSMUST00000022856.15
ENSMUST00000100775.10 ENSMUST00000169519.8 |
Rad1
|
RAD1 checkpoint DNA exonuclease |
| chr3_-_103716593 | 0.03 |
ENSMUST00000063502.13
ENSMUST00000106832.2 ENSMUST00000106834.8 ENSMUST00000029435.15 |
Dclre1b
|
DNA cross-link repair 1B |
| chr11_+_40624763 | 0.03 |
ENSMUST00000127382.2
|
Nudcd2
|
NudC domain containing 2 |
| chr5_+_145020640 | 0.03 |
ENSMUST00000031625.15
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr15_-_66673425 | 0.03 |
ENSMUST00000168589.8
|
Sla
|
src-like adaptor |
| chr11_-_116089866 | 0.03 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr12_+_116011332 | 0.03 |
ENSMUST00000073551.6
ENSMUST00000183125.2 |
Zfp386
|
zinc finger protein 386 (Kruppel-like) |
| chr12_-_72455708 | 0.03 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr1_+_171723231 | 0.03 |
ENSMUST00000097466.3
|
Gm10521
|
predicted gene 10521 |
| chr11_-_3599673 | 0.03 |
ENSMUST00000193809.2
|
Tug1
|
taurine upregulated gene 1 |
| chr7_-_119461027 | 0.03 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chrX_-_48980360 | 0.03 |
ENSMUST00000217355.3
|
Olfr1322
|
olfactory receptor 1322 |
| chr1_-_167112784 | 0.03 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr15_-_10485976 | 0.02 |
ENSMUST00000169050.8
ENSMUST00000022855.12 |
Brix1
|
BRX1, biogenesis of ribosomes |
| chr1_-_172418058 | 0.02 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8 |
| chr6_-_124689094 | 0.02 |
ENSMUST00000004379.8
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr1_+_75119472 | 0.02 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr2_+_91541197 | 0.02 |
ENSMUST00000128140.2
ENSMUST00000140183.2 |
Harbi1
|
harbinger transposase derived 1 |
| chr3_-_107838895 | 0.02 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr14_-_55101505 | 0.02 |
ENSMUST00000142283.4
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr7_-_126807581 | 0.02 |
ENSMUST00000120705.3
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr19_-_43928583 | 0.02 |
ENSMUST00000212396.2
|
Dnmbp
|
dynamin binding protein |
| chr17_-_52117894 | 0.02 |
ENSMUST00000176669.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr13_+_21679387 | 0.02 |
ENSMUST00000104942.2
|
AK157302
|
cDNA sequence AK157302 |
| chr14_-_40730180 | 0.02 |
ENSMUST00000136661.8
|
Prxl2a
|
peroxiredoxin like 2A |
| chr15_+_58287305 | 0.02 |
ENSMUST00000037270.5
|
Fam91a1
|
family with sequence similarity 91, member A1 |
| chr6_-_30873669 | 0.02 |
ENSMUST00000048774.13
ENSMUST00000166192.7 |
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr11_-_100418688 | 0.02 |
ENSMUST00000107385.2
|
Acly
|
ATP citrate lyase |
| chr7_+_126895531 | 0.02 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr16_-_21814289 | 0.02 |
ENSMUST00000060673.8
|
Liph
|
lipase, member H |
| chr11_-_116089595 | 0.02 |
ENSMUST00000072948.11
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr7_+_28393419 | 0.02 |
ENSMUST00000108280.2
|
Fbxo27
|
F-box protein 27 |
| chr2_+_57887896 | 0.02 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr3_+_137573436 | 0.01 |
ENSMUST00000090178.10
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr11_-_115519086 | 0.01 |
ENSMUST00000178003.8
|
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr10_-_116808514 | 0.01 |
ENSMUST00000092165.5
|
Gm10271
|
predicted gene 10271 |
| chr7_+_139900771 | 0.01 |
ENSMUST00000214594.2
|
Olfr525
|
olfactory receptor 525 |
| chr16_-_44379226 | 0.01 |
ENSMUST00000114634.3
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| chr11_-_119907884 | 0.01 |
ENSMUST00000132575.8
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr14_-_51134930 | 0.01 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr15_-_5137951 | 0.01 |
ENSMUST00000141020.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr10_-_128237087 | 0.01 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr5_+_4073343 | 0.01 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr17_+_47680097 | 0.01 |
ENSMUST00000060752.13
ENSMUST00000119841.8 |
Mrps10
|
mitochondrial ribosomal protein S10 |
| chr7_-_99770280 | 0.01 |
ENSMUST00000208184.2
|
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr8_-_3674993 | 0.01 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr12_+_71184614 | 0.01 |
ENSMUST00000045907.16
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr2_-_91540864 | 0.01 |
ENSMUST00000028678.9
ENSMUST00000076803.12 |
Atg13
|
autophagy related 13 |
| chr2_+_86832276 | 0.01 |
ENSMUST00000055129.6
|
Olfr1102
|
olfactory receptor 1102 |
| chr9_+_65368207 | 0.01 |
ENSMUST00000034955.8
ENSMUST00000213957.2 |
Spg21
|
SPG21, maspardin |
| chr4_-_33189390 | 0.01 |
ENSMUST00000098181.9
|
Pm20d2
|
peptidase M20 domain containing 2 |
| chr4_-_20778847 | 0.01 |
ENSMUST00000102998.4
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr2_-_5867734 | 0.01 |
ENSMUST00000071016.3
|
Gm13199
|
predicted gene 13199 |
| chrX_+_102465616 | 0.01 |
ENSMUST00000182089.2
|
Gm26992
|
predicted gene, 26992 |
| chr1_+_86631510 | 0.01 |
ENSMUST00000168237.8
ENSMUST00000065694.8 |
Dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr2_+_155593030 | 0.00 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr1_-_120432477 | 0.00 |
ENSMUST00000186432.3
|
Marco
|
macrophage receptor with collagenous structure |
| chr2_+_158344553 | 0.00 |
ENSMUST00000109484.2
|
Adig
|
adipogenin |
| chr19_-_47680528 | 0.00 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr8_-_3675274 | 0.00 |
ENSMUST00000004749.7
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr2_+_158344532 | 0.00 |
ENSMUST00000059889.4
|
Adig
|
adipogenin |
| chr15_-_97629209 | 0.00 |
ENSMUST00000100249.10
|
Endou
|
endonuclease, polyU-specific |
| chr14_-_51134906 | 0.00 |
ENSMUST00000170855.2
|
Klhl33
|
kelch-like 33 |
| chr11_+_95603494 | 0.00 |
ENSMUST00000107717.8
|
Zfp652
|
zinc finger protein 652 |
| chr15_-_5137975 | 0.00 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chr6_+_29361408 | 0.00 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr4_-_116228921 | 0.00 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr1_-_180021039 | 0.00 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr1_+_75119809 | 0.00 |
ENSMUST00000186037.7
ENSMUST00000187901.2 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr7_-_141729738 | 0.00 |
ENSMUST00000190456.2
ENSMUST00000067978.6 |
Krtap5-2
|
keratin associated protein 5-2 |
| chr3_+_90507510 | 0.00 |
ENSMUST00000001047.8
|
S100a3
|
S100 calcium binding protein A3 |
| chr7_+_126808016 | 0.00 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr6_+_70332836 | 0.00 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr8_-_107792264 | 0.00 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr5_+_90708962 | 0.00 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr7_+_126895463 | 0.00 |
ENSMUST00000106306.9
ENSMUST00000120857.8 |
Itgal
|
integrin alpha L |
| chr7_+_126810780 | 0.00 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr18_-_39000056 | 0.00 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr5_+_88635834 | 0.00 |
ENSMUST00000199104.5
ENSMUST00000031222.9 |
Enam
|
enamelin |
| chr14_+_53100756 | 0.00 |
ENSMUST00000103616.5
ENSMUST00000186370.2 |
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr11_-_100418717 | 0.00 |
ENSMUST00000107389.8
ENSMUST00000007131.16 |
Acly
|
ATP citrate lyase |
| chr3_-_92493507 | 0.00 |
ENSMUST00000194965.6
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rorc_Nr1d1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.0 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.2 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.0 | 0.1 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.0 | 0.1 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of eosinophil activation(GO:1902567) negative regulation of activation of JAK2 kinase activity(GO:1902569) negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.0 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.0 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |