Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
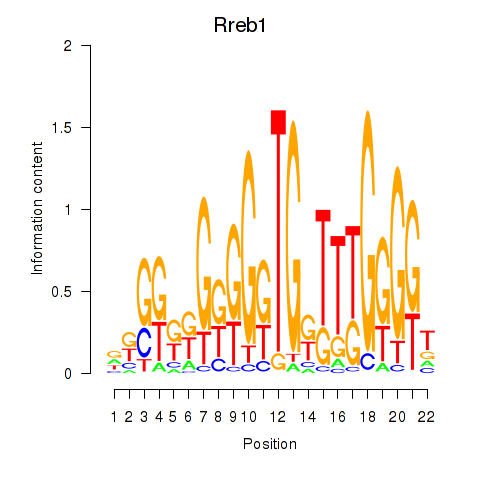
Results for Rreb1
Z-value: 0.31
Transcription factors associated with Rreb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rreb1
|
ENSMUSG00000039087.18 | ras responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rreb1 | mm39_v1_chr13_+_38010203_38010291 | 0.85 | 6.6e-02 | Click! |
Activity profile of Rreb1 motif
Sorted Z-values of Rreb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_109678685 | 0.32 |
ENSMUST00000112022.5
|
Camp
|
cathelicidin antimicrobial peptide |
| chr1_-_72914036 | 0.22 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr4_-_62126662 | 0.19 |
ENSMUST00000222050.2
ENSMUST00000068822.4 |
Zfp37
|
zinc finger protein 37 |
| chr6_-_67316645 | 0.14 |
ENSMUST00000117441.8
|
Il12rb2
|
interleukin 12 receptor, beta 2 |
| chrX_-_71318353 | 0.13 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr14_+_69846927 | 0.13 |
ENSMUST00000100420.4
|
Loxl2
|
lysyl oxidase-like 2 |
| chr14_+_69846517 | 0.12 |
ENSMUST00000022660.14
|
Loxl2
|
lysyl oxidase-like 2 |
| chr5_-_108822619 | 0.12 |
ENSMUST00000119270.2
ENSMUST00000163328.8 ENSMUST00000136227.2 |
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr13_-_42000958 | 0.11 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr17_+_34124078 | 0.11 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chrX_+_7744535 | 0.11 |
ENSMUST00000033495.15
|
Pim2
|
proviral integration site 2 |
| chr9_+_38407097 | 0.10 |
ENSMUST00000214003.3
ENSMUST00000214264.3 |
Olfr907
|
olfactory receptor 907 |
| chr10_+_79984097 | 0.10 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chrX_+_149330371 | 0.10 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr17_-_32074754 | 0.10 |
ENSMUST00000024839.6
|
Sik1
|
salt inducible kinase 1 |
| chr4_-_3938352 | 0.10 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chrX_-_104972150 | 0.09 |
ENSMUST00000101305.9
|
Atrx
|
ATRX, chromatin remodeler |
| chr17_-_81977590 | 0.09 |
ENSMUST00000234923.2
|
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr10_+_4296806 | 0.09 |
ENSMUST00000216139.3
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr17_-_56019571 | 0.09 |
ENSMUST00000086876.7
ENSMUST00000233557.2 ENSMUST00000233972.2 |
Pot1b
|
protection of telomeres 1B |
| chr19_+_10502679 | 0.09 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr11_-_65636651 | 0.08 |
ENSMUST00000138093.2
|
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr8_+_108020132 | 0.08 |
ENSMUST00000151114.8
ENSMUST00000125721.8 ENSMUST00000075922.11 |
Nfat5
|
nuclear factor of activated T cells 5 |
| chr11_+_98828495 | 0.08 |
ENSMUST00000107475.9
ENSMUST00000068133.10 |
Rara
|
retinoic acid receptor, alpha |
| chr8_+_108020092 | 0.08 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr17_-_29768586 | 0.08 |
ENSMUST00000234305.2
ENSMUST00000234648.2 ENSMUST00000234979.2 |
Gm17657
Tmem217
|
predicted gene, 17657 transmembrane protein 217 |
| chr1_+_58752415 | 0.08 |
ENSMUST00000114309.8
ENSMUST00000069333.8 |
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chr10_-_128953730 | 0.08 |
ENSMUST00000215453.2
ENSMUST00000216906.2 |
Olfr769
|
olfactory receptor 769 |
| chr13_-_93810808 | 0.08 |
ENSMUST00000015941.8
|
Bhmt2
|
betaine-homocysteine methyltransferase 2 |
| chr8_+_127025265 | 0.08 |
ENSMUST00000108759.3
|
Slc35f3
|
solute carrier family 35, member F3 |
| chr18_-_38131766 | 0.08 |
ENSMUST00000236588.2
ENSMUST00000237272.2 ENSMUST00000236134.2 |
Arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr12_+_84161095 | 0.07 |
ENSMUST00000123491.8
ENSMUST00000046340.9 ENSMUST00000136159.2 |
Dnal1
|
dynein, axonemal, light chain 1 |
| chrX_+_137815171 | 0.07 |
ENSMUST00000064937.14
ENSMUST00000113052.8 |
Nrk
|
Nik related kinase |
| chr5_-_124563611 | 0.07 |
ENSMUST00000198420.5
|
Sbno1
|
strawberry notch 1 |
| chr12_-_84240781 | 0.07 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr14_-_7666348 | 0.07 |
ENSMUST00000224491.2
|
Nek10
|
NIMA (never in mitosis gene a)- related kinase 10 |
| chr19_-_10502468 | 0.07 |
ENSMUST00000025570.8
ENSMUST00000236455.2 |
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr16_-_36228798 | 0.07 |
ENSMUST00000023619.8
|
Stfa2
|
stefin A2 |
| chr2_-_154214622 | 0.07 |
ENSMUST00000028990.10
ENSMUST00000109730.3 |
Cdk5rap1
|
CDK5 regulatory subunit associated protein 1 |
| chr2_-_33261498 | 0.07 |
ENSMUST00000113165.8
|
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr9_+_22136998 | 0.07 |
ENSMUST00000215902.2
|
Zfp809
|
zinc finger protein 809 |
| chr5_+_147894121 | 0.07 |
ENSMUST00000085558.11
ENSMUST00000129092.2 |
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr9_+_106247943 | 0.07 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr9_-_48252176 | 0.07 |
ENSMUST00000034527.14
|
Nxpe2
|
neurexophilin and PC-esterase domain family, member 2 |
| chr9_+_118881838 | 0.07 |
ENSMUST00000051386.13
ENSMUST00000074734.13 |
Vill
|
villin-like |
| chr19_+_10502612 | 0.06 |
ENSMUST00000237321.2
ENSMUST00000038379.5 |
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr4_-_15149755 | 0.06 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr6_-_83433357 | 0.06 |
ENSMUST00000186548.7
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr7_-_46608193 | 0.06 |
ENSMUST00000094398.13
|
Uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr5_-_124563636 | 0.06 |
ENSMUST00000196711.5
ENSMUST00000200474.5 ENSMUST00000199808.5 |
Sbno1
|
strawberry notch 1 |
| chr12_+_102249375 | 0.06 |
ENSMUST00000101114.11
ENSMUST00000150795.8 |
Rin3
|
Ras and Rab interactor 3 |
| chr5_-_114131779 | 0.06 |
ENSMUST00000112298.10
|
Ssh1
|
slingshot protein phosphatase 1 |
| chr7_-_105282687 | 0.06 |
ENSMUST00000147044.4
ENSMUST00000106791.8 ENSMUST00000153371.9 ENSMUST00000106789.8 |
Trim3
|
tripartite motif-containing 3 |
| chr3_-_32670628 | 0.06 |
ENSMUST00000193050.2
ENSMUST00000108234.8 ENSMUST00000155737.8 |
Gnb4
|
guanine nucleotide binding protein (G protein), beta 4 |
| chr4_+_97665992 | 0.06 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr14_-_100521888 | 0.06 |
ENSMUST00000226774.2
|
Klf12
|
Kruppel-like factor 12 |
| chr7_-_23510068 | 0.06 |
ENSMUST00000228383.2
|
Vmn1r175
|
vomeronasal 1 receptor 175 |
| chr12_-_112766266 | 0.06 |
ENSMUST00000239525.1
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr5_-_124564014 | 0.06 |
ENSMUST00000196329.5
ENSMUST00000196644.5 |
Sbno1
|
strawberry notch 1 |
| chr7_-_29426425 | 0.06 |
ENSMUST00000061193.4
|
Catsperg2
|
cation channel sperm associated auxiliary subunit gamma 2 |
| chr1_-_123973223 | 0.06 |
ENSMUST00000112606.8
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr17_+_43878989 | 0.06 |
ENSMUST00000167214.8
ENSMUST00000024706.12 |
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr15_+_84565174 | 0.05 |
ENSMUST00000065499.5
|
Prr5
|
proline rich 5 (renal) |
| chr16_+_75389732 | 0.05 |
ENSMUST00000046378.14
ENSMUST00000114249.8 ENSMUST00000114253.2 |
Rbm11
|
RNA binding motif protein 11 |
| chr19_+_8568618 | 0.05 |
ENSMUST00000170817.2
ENSMUST00000010251.11 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr18_+_82929451 | 0.05 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr5_+_140721281 | 0.05 |
ENSMUST00000120630.3
|
Amz1
|
archaelysin family metallopeptidase 1 |
| chr7_-_105282751 | 0.05 |
ENSMUST00000057525.14
|
Trim3
|
tripartite motif-containing 3 |
| chr7_+_24596806 | 0.05 |
ENSMUST00000003469.8
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr2_-_31006302 | 0.05 |
ENSMUST00000149196.2
|
Fnbp1
|
formin binding protein 1 |
| chr5_+_123390149 | 0.05 |
ENSMUST00000121964.8
|
Wdr66
|
WD repeat domain 66 |
| chr1_-_132953068 | 0.05 |
ENSMUST00000186617.7
ENSMUST00000067429.10 ENSMUST00000067398.13 ENSMUST00000188090.7 |
Mdm4
|
transformed mouse 3T3 cell double minute 4 |
| chr17_+_29587930 | 0.05 |
ENSMUST00000137644.3
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr4_-_116001737 | 0.05 |
ENSMUST00000030469.5
|
Lurap1
|
leucine rich adaptor protein 1 |
| chr11_-_94133527 | 0.05 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr11_+_115802828 | 0.05 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr15_-_74713458 | 0.05 |
ENSMUST00000170259.3
|
Cyp11b1
|
cytochrome P450, family 11, subfamily b, polypeptide 1 |
| chr4_-_15149051 | 0.05 |
ENSMUST00000041606.14
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr6_-_97464761 | 0.05 |
ENSMUST00000032146.14
|
Frmd4b
|
FERM domain containing 4B |
| chr11_-_103247150 | 0.05 |
ENSMUST00000136491.3
ENSMUST00000107023.3 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr17_-_33007238 | 0.05 |
ENSMUST00000159086.10
|
Zfp871
|
zinc finger protein 871 |
| chr2_+_11710523 | 0.05 |
ENSMUST00000138856.2
ENSMUST00000078834.12 ENSMUST00000114834.10 ENSMUST00000114833.10 ENSMUST00000114831.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr1_-_138775317 | 0.05 |
ENSMUST00000093486.10
ENSMUST00000046870.13 |
Lhx9
|
LIM homeobox protein 9 |
| chr10_-_7423606 | 0.05 |
ENSMUST00000177585.9
|
Ulbp1
|
UL16 binding protein 1 |
| chr18_+_82932747 | 0.05 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr11_+_79482005 | 0.04 |
ENSMUST00000017783.13
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chr17_+_34848507 | 0.04 |
ENSMUST00000015620.7
|
Prrt1
|
proline-rich transmembrane protein 1 |
| chr4_-_41098174 | 0.04 |
ENSMUST00000055327.8
|
Aqp3
|
aquaporin 3 |
| chr16_+_62674661 | 0.04 |
ENSMUST00000023629.9
|
Pros1
|
protein S (alpha) |
| chr10_-_7423341 | 0.04 |
ENSMUST00000169796.4
ENSMUST00000218087.2 |
Ulbp1
|
UL16 binding protein 1 |
| chr6_+_48715971 | 0.04 |
ENSMUST00000054368.7
ENSMUST00000140054.3 ENSMUST00000204168.2 ENSMUST00000204408.2 |
Gimap1
Gm28053
|
GTPase, IMAP family member 1 predicted gene, 28053 |
| chr19_-_4241034 | 0.04 |
ENSMUST00000237495.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr2_-_33261411 | 0.04 |
ENSMUST00000131298.7
ENSMUST00000091039.5 ENSMUST00000042615.13 |
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr9_+_121232480 | 0.04 |
ENSMUST00000210351.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chr2_-_63014622 | 0.04 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr8_-_73175283 | 0.04 |
ENSMUST00000212121.2
ENSMUST00000212590.2 ENSMUST00000163643.3 |
Eps15l1
|
epidermal growth factor receptor pathway substrate 15-like 1 |
| chr8_-_3744167 | 0.04 |
ENSMUST00000005678.6
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide |
| chr18_+_31937129 | 0.04 |
ENSMUST00000082319.15
ENSMUST00000234957.2 ENSMUST00000025264.8 ENSMUST00000234344.2 |
Wdr33
|
WD repeat domain 33 |
| chr15_-_89033761 | 0.04 |
ENSMUST00000088823.5
|
Mapk11
|
mitogen-activated protein kinase 11 |
| chr7_+_4693759 | 0.04 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr9_+_98305014 | 0.04 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chr4_-_117740624 | 0.04 |
ENSMUST00000030266.12
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr17_+_47696329 | 0.04 |
ENSMUST00000145462.2
|
Guca1b
|
guanylate cyclase activator 1B |
| chr6_+_37507108 | 0.04 |
ENSMUST00000040987.11
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr15_-_85387428 | 0.04 |
ENSMUST00000178942.2
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chr17_-_36331416 | 0.04 |
ENSMUST00000174063.2
ENSMUST00000113760.10 |
H2-T24
|
histocompatibility 2, T region locus 24 |
| chr1_-_87936242 | 0.04 |
ENSMUST00000187758.7
ENSMUST00000040783.11 |
Usp40
|
ubiquitin specific peptidase 40 |
| chr9_-_100453102 | 0.04 |
ENSMUST00000093792.4
|
Slc35g2
|
solute carrier family 35, member G2 |
| chr13_+_94495457 | 0.04 |
ENSMUST00000022196.5
|
Ap3b1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr6_-_83436066 | 0.04 |
ENSMUST00000190295.2
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr6_+_83214357 | 0.04 |
ENSMUST00000039212.8
ENSMUST00000113899.8 |
Slc4a5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5 |
| chr12_+_12312135 | 0.04 |
ENSMUST00000069066.14
|
Cyria
|
CYFIP related Rac1 interactor A |
| chr11_-_6469494 | 0.04 |
ENSMUST00000134489.2
|
Myo1g
|
myosin IG |
| chr16_+_51852435 | 0.04 |
ENSMUST00000227879.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr7_+_28416270 | 0.04 |
ENSMUST00000108279.9
|
Fbxo17
|
F-box protein 17 |
| chr17_-_56583715 | 0.04 |
ENSMUST00000058136.9
|
Ticam1
|
toll-like receptor adaptor molecule 1 |
| chr19_-_4240984 | 0.04 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr5_+_136023649 | 0.04 |
ENSMUST00000111142.9
ENSMUST00000111145.10 ENSMUST00000111144.8 ENSMUST00000199239.5 ENSMUST00000005072.10 ENSMUST00000130345.2 |
Dtx2
|
deltex 2, E3 ubiquitin ligase |
| chr4_+_155648256 | 0.04 |
ENSMUST00000143840.2
ENSMUST00000146080.8 |
Nadk
|
NAD kinase |
| chr5_+_114845821 | 0.03 |
ENSMUST00000094441.11
|
Tchp
|
trichoplein, keratin filament binding |
| chr5_-_147662798 | 0.03 |
ENSMUST00000110529.6
ENSMUST00000031653.12 |
Flt1
|
FMS-like tyrosine kinase 1 |
| chr7_-_138447933 | 0.03 |
ENSMUST00000118810.2
ENSMUST00000075667.5 ENSMUST00000119664.2 |
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr3_+_40594899 | 0.03 |
ENSMUST00000091186.7
|
Intu
|
inturned planar cell polarity protein |
| chr9_-_65815958 | 0.03 |
ENSMUST00000119245.8
ENSMUST00000134338.8 ENSMUST00000179395.8 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr10_+_86136236 | 0.03 |
ENSMUST00000020234.14
|
Timp3
|
tissue inhibitor of metalloproteinase 3 |
| chr1_-_181847492 | 0.03 |
ENSMUST00000177811.8
ENSMUST00000111025.8 ENSMUST00000111024.10 |
Enah
|
ENAH actin regulator |
| chr14_+_32043944 | 0.03 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr1_-_36312482 | 0.03 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr5_-_121975676 | 0.03 |
ENSMUST00000197892.3
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr19_+_3817953 | 0.03 |
ENSMUST00000113970.8
|
Kmt5b
|
lysine methyltransferase 5B |
| chr9_+_65816370 | 0.03 |
ENSMUST00000206594.2
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr4_-_132191181 | 0.03 |
ENSMUST00000102567.4
|
Med18
|
mediator complex subunit 18 |
| chr17_-_26063488 | 0.03 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr9_+_100479732 | 0.03 |
ENSMUST00000124487.8
|
Stag1
|
stromal antigen 1 |
| chr2_+_181023028 | 0.03 |
ENSMUST00000048077.12
|
Lime1
|
Lck interacting transmembrane adaptor 1 |
| chr12_-_16639756 | 0.03 |
ENSMUST00000222989.2
ENSMUST00000067124.6 ENSMUST00000221230.2 |
Lpin1
|
lipin 1 |
| chr4_-_43010225 | 0.03 |
ENSMUST00000030165.5
|
Fancg
|
Fanconi anemia, complementation group G |
| chr4_+_62204678 | 0.03 |
ENSMUST00000084530.9
|
Slc31a2
|
solute carrier family 31, member 2 |
| chr17_-_30795136 | 0.03 |
ENSMUST00000079924.8
ENSMUST00000236584.2 ENSMUST00000236825.2 ENSMUST00000235587.2 |
Btbd9
|
BTB (POZ) domain containing 9 |
| chr7_+_4693603 | 0.03 |
ENSMUST00000120836.8
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr11_-_114851243 | 0.03 |
ENSMUST00000092466.13
ENSMUST00000061637.4 |
Cd300c
|
CD300C molecule |
| chr17_+_44263890 | 0.03 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr13_-_63579497 | 0.03 |
ENSMUST00000160931.2
ENSMUST00000099444.10 ENSMUST00000220684.2 ENSMUST00000161977.8 ENSMUST00000163091.8 |
Fancc
|
Fanconi anemia, complementation group C |
| chr13_-_41513215 | 0.03 |
ENSMUST00000224803.2
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9 |
| chr7_+_101583283 | 0.03 |
ENSMUST00000209639.2
ENSMUST00000210679.2 |
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr5_+_150042092 | 0.03 |
ENSMUST00000200960.4
ENSMUST00000202530.4 |
Fry
|
FRY microtubule binding protein |
| chr17_-_45047521 | 0.03 |
ENSMUST00000113572.9
|
Runx2
|
runt related transcription factor 2 |
| chr15_+_99600475 | 0.03 |
ENSMUST00000228984.2
ENSMUST00000229845.2 |
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr1_-_192955407 | 0.03 |
ENSMUST00000009777.4
|
G0s2
|
G0/G1 switch gene 2 |
| chr8_+_70261323 | 0.03 |
ENSMUST00000036074.15
ENSMUST00000123453.2 |
Gmip
|
Gem-interacting protein |
| chr8_+_10056654 | 0.03 |
ENSMUST00000033892.9
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr13_-_63579551 | 0.03 |
ENSMUST00000073029.13
|
Fancc
|
Fanconi anemia, complementation group C |
| chr5_-_114796425 | 0.03 |
ENSMUST00000112225.8
ENSMUST00000071968.9 |
Trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr7_+_103199575 | 0.03 |
ENSMUST00000106888.3
|
Olfr613
|
olfactory receptor 613 |
| chr19_+_23881821 | 0.03 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr7_+_140547134 | 0.03 |
ENSMUST00000106042.9
|
Ifitm1
|
interferon induced transmembrane protein 1 |
| chr17_+_45866618 | 0.03 |
ENSMUST00000024742.9
ENSMUST00000233929.2 |
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon |
| chr9_+_65816206 | 0.03 |
ENSMUST00000205379.2
ENSMUST00000206048.2 ENSMUST00000034949.10 ENSMUST00000154589.2 |
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr6_+_17307272 | 0.03 |
ENSMUST00000115454.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr8_-_29709652 | 0.03 |
ENSMUST00000168630.4
|
Unc5d
|
unc-5 netrin receptor D |
| chr5_-_114131934 | 0.03 |
ENSMUST00000159592.8
|
Ssh1
|
slingshot protein phosphatase 1 |
| chr7_-_97827461 | 0.03 |
ENSMUST00000040971.14
|
Capn5
|
calpain 5 |
| chr8_-_94738748 | 0.03 |
ENSMUST00000143265.2
|
Amfr
|
autocrine motility factor receptor |
| chr7_-_126194097 | 0.03 |
ENSMUST00000058429.6
|
Il27
|
interleukin 27 |
| chr7_+_16576821 | 0.03 |
ENSMUST00000168093.9
|
Prkd2
|
protein kinase D2 |
| chr2_+_26518456 | 0.03 |
ENSMUST00000074240.4
|
Dipk1b
|
divergent protein kinase domain 1B |
| chr17_-_48189815 | 0.03 |
ENSMUST00000154108.2
|
Foxp4
|
forkhead box P4 |
| chr6_+_65648574 | 0.03 |
ENSMUST00000054351.6
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr2_+_122607297 | 0.03 |
ENSMUST00000124460.2
ENSMUST00000147475.2 |
Sqor
|
sulfide quinone oxidoreductase |
| chr15_-_81845019 | 0.03 |
ENSMUST00000230229.2
|
Pmm1
|
phosphomannomutase 1 |
| chr6_-_115229128 | 0.03 |
ENSMUST00000032462.9
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr6_+_68414401 | 0.03 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr12_+_83678987 | 0.03 |
ENSMUST00000048155.16
ENSMUST00000182618.8 ENSMUST00000183154.8 ENSMUST00000182036.8 ENSMUST00000182347.8 |
Rbm25
|
RNA binding motif protein 25 |
| chr7_+_16576978 | 0.03 |
ENSMUST00000086104.6
|
Prkd2
|
protein kinase D2 |
| chr1_+_59952131 | 0.03 |
ENSMUST00000036540.12
|
Fam117b
|
family with sequence similarity 117, member B |
| chr5_+_125080504 | 0.03 |
ENSMUST00000197746.2
|
Rflna
|
refilin A |
| chr4_+_43441939 | 0.02 |
ENSMUST00000060864.13
|
Tesk1
|
testis specific protein kinase 1 |
| chr5_-_120726721 | 0.02 |
ENSMUST00000046426.10
|
Tpcn1
|
two pore channel 1 |
| chr6_-_99412306 | 0.02 |
ENSMUST00000113322.9
ENSMUST00000176850.8 ENSMUST00000176632.8 |
Foxp1
|
forkhead box P1 |
| chr6_+_4903299 | 0.02 |
ENSMUST00000035813.9
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr2_+_84508734 | 0.02 |
ENSMUST00000102645.4
|
Med19
|
mediator complex subunit 19 |
| chr2_-_9883391 | 0.02 |
ENSMUST00000102976.4
|
Gata3
|
GATA binding protein 3 |
| chr19_-_10502546 | 0.02 |
ENSMUST00000237827.2
|
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr14_-_60324265 | 0.02 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr7_+_5059703 | 0.02 |
ENSMUST00000208042.2
ENSMUST00000207974.2 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr10_-_7831657 | 0.02 |
ENSMUST00000147938.2
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr3_+_142236086 | 0.02 |
ENSMUST00000171263.8
ENSMUST00000045097.11 |
Gbp7
|
guanylate binding protein 7 |
| chr5_-_74189898 | 0.02 |
ENSMUST00000152408.3
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr19_+_3818112 | 0.02 |
ENSMUST00000005518.16
ENSMUST00000237440.2 ENSMUST00000152935.8 ENSMUST00000176262.8 ENSMUST00000176407.8 ENSMUST00000176926.8 ENSMUST00000176512.8 |
Kmt5b
|
lysine methyltransferase 5B |
| chr4_-_141660390 | 0.02 |
ENSMUST00000036701.8
|
Fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr6_+_48624295 | 0.02 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
| chr7_+_138794577 | 0.02 |
ENSMUST00000135509.2
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr7_+_44240310 | 0.02 |
ENSMUST00000107906.6
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr7_+_45135682 | 0.02 |
ENSMUST00000210813.2
ENSMUST00000107759.10 |
Tulp2
|
tubby-like protein 2 |
| chr1_-_166237341 | 0.02 |
ENSMUST00000135673.8
ENSMUST00000169324.8 ENSMUST00000128861.3 |
Pogk
|
pogo transposable element with KRAB domain |
| chr3_+_130904000 | 0.02 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr19_-_4333061 | 0.02 |
ENSMUST00000167215.2
ENSMUST00000056888.13 |
Ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr6_-_129308748 | 0.02 |
ENSMUST00000051283.8
|
Clec2m
|
C-type lectin domain family 2, member m |
| chr1_-_84262274 | 0.02 |
ENSMUST00000177458.2
ENSMUST00000168574.9 |
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr18_-_36812406 | 0.02 |
ENSMUST00000001415.9
|
Apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rreb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.1 | GO:1901740 | skeletal muscle atrophy(GO:0014732) negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 0.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.0 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |