Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
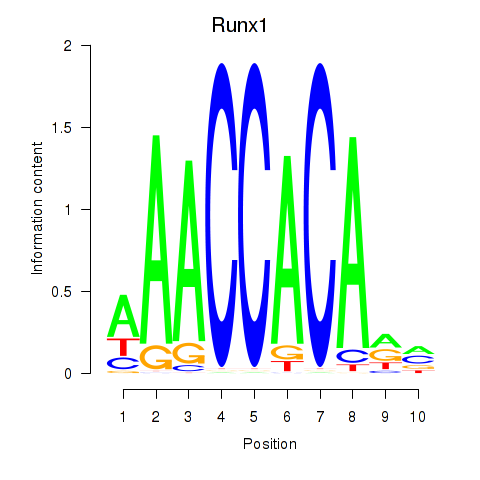
Results for Runx1
Z-value: 0.44
Transcription factors associated with Runx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx1
|
ENSMUSG00000022952.18 | runt related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx1 | mm39_v1_chr16_-_92494203_92494235 | -0.67 | 2.1e-01 | Click! |
Activity profile of Runx1 motif
Sorted Z-values of Runx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_140111018 | 0.46 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr1_-_140111138 | 0.44 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr5_-_107873883 | 0.37 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr2_+_164790139 | 0.23 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr16_-_95387444 | 0.22 |
ENSMUST00000233269.2
|
Erg
|
ETS transcription factor |
| chr9_-_43151179 | 0.20 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr4_+_102617495 | 0.19 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr12_+_10440755 | 0.18 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr11_+_87684299 | 0.17 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr11_-_83540175 | 0.17 |
ENSMUST00000001008.6
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr17_+_26882171 | 0.17 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr13_-_113237505 | 0.17 |
ENSMUST00000224282.2
ENSMUST00000023897.7 |
Gzma
|
granzyme A |
| chr14_-_56499690 | 0.15 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr4_+_102848981 | 0.14 |
ENSMUST00000140654.9
|
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr6_+_17749169 | 0.14 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr19_-_10857734 | 0.14 |
ENSMUST00000133303.8
|
Tmem109
|
transmembrane protein 109 |
| chr1_+_60785517 | 0.13 |
ENSMUST00000027165.3
|
Cd28
|
CD28 antigen |
| chr7_+_44221791 | 0.13 |
ENSMUST00000002274.10
|
Napsa
|
napsin A aspartic peptidase |
| chr11_+_87684548 | 0.13 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr14_-_31362835 | 0.13 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr2_+_172235820 | 0.12 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr5_+_104582978 | 0.12 |
ENSMUST00000086833.13
ENSMUST00000031243.15 ENSMUST00000112748.8 ENSMUST00000112746.8 ENSMUST00000145084.8 ENSMUST00000132457.8 |
Spp1
|
secreted phosphoprotein 1 |
| chr17_-_37269153 | 0.12 |
ENSMUST00000172823.2
|
Polr1h
|
RNA polymerase I subunit H |
| chr9_+_113641615 | 0.12 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr11_-_69786324 | 0.12 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr6_-_11907392 | 0.11 |
ENSMUST00000204084.3
ENSMUST00000031637.8 ENSMUST00000204978.3 ENSMUST00000204714.2 |
Ndufa4
|
Ndufa4, mitochondrial complex associated |
| chr2_+_68966125 | 0.11 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chr13_+_76727787 | 0.11 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr9_-_103569984 | 0.11 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr1_+_88015524 | 0.10 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr4_-_152122891 | 0.09 |
ENSMUST00000030792.2
|
Tas1r1
|
taste receptor, type 1, member 1 |
| chr10_-_35587888 | 0.09 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr4_-_133600308 | 0.09 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_-_72106418 | 0.09 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr1_-_192946359 | 0.09 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr19_+_4264470 | 0.09 |
ENSMUST00000237171.2
|
Gm45928
|
predicted gene, 45928 |
| chr10_-_40178182 | 0.09 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr11_-_115426618 | 0.09 |
ENSMUST00000121185.8
ENSMUST00000117589.8 |
Sumo2
|
small ubiquitin-like modifier 2 |
| chr5_-_140368482 | 0.08 |
ENSMUST00000196566.5
|
Snx8
|
sorting nexin 8 |
| chr1_+_88022776 | 0.08 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr11_-_100986192 | 0.08 |
ENSMUST00000019447.15
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr7_-_45083688 | 0.08 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr16_+_91188609 | 0.08 |
ENSMUST00000160764.2
|
Gm21970
|
predicted gene 21970 |
| chr15_-_38519499 | 0.08 |
ENSMUST00000110329.8
ENSMUST00000065308.13 |
Azin1
|
antizyme inhibitor 1 |
| chr11_-_46280298 | 0.08 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr17_-_37269330 | 0.08 |
ENSMUST00000113669.9
|
Polr1h
|
RNA polymerase I subunit H |
| chr19_+_6111204 | 0.07 |
ENSMUST00000162726.5
|
Znhit2
|
zinc finger, HIT domain containing 2 |
| chr15_-_103163860 | 0.07 |
ENSMUST00000075192.13
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr13_+_54937312 | 0.07 |
ENSMUST00000163915.8
ENSMUST00000099503.10 ENSMUST00000171859.8 |
Tspan17
|
tetraspanin 17 |
| chr11_-_46280336 | 0.07 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr19_+_53665719 | 0.07 |
ENSMUST00000164202.9
|
Rbm20
|
RNA binding motif protein 20 |
| chr2_+_30331839 | 0.07 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr3_-_107667499 | 0.07 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr17_+_37269513 | 0.07 |
ENSMUST00000173814.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr17_-_37269425 | 0.06 |
ENSMUST00000172518.8
|
Polr1h
|
RNA polymerase I subunit H |
| chr19_+_8816663 | 0.06 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr10_-_5144699 | 0.06 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr14_-_31362909 | 0.06 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr2_-_20948230 | 0.06 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr17_+_31427023 | 0.06 |
ENSMUST00000173776.2
ENSMUST00000048656.15 |
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chrX_-_12026594 | 0.06 |
ENSMUST00000043441.13
|
Bcor
|
BCL6 interacting corepressor |
| chr1_+_40554513 | 0.06 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr7_+_133311062 | 0.05 |
ENSMUST00000033282.5
|
Bccip
|
BRCA2 and CDKN1A interacting protein |
| chrX_-_100777806 | 0.05 |
ENSMUST00000056614.7
|
Cxcr3
|
chemokine (C-X-C motif) receptor 3 |
| chr9_-_56151334 | 0.05 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr11_-_69576363 | 0.05 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr19_-_10807409 | 0.05 |
ENSMUST00000080292.12
|
Cd6
|
CD6 antigen |
| chr17_+_37269468 | 0.05 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr1_-_164763091 | 0.05 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr3_-_89000591 | 0.05 |
ENSMUST00000090929.12
ENSMUST00000052539.13 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr4_-_129467430 | 0.05 |
ENSMUST00000102596.8
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr8_+_77628916 | 0.05 |
ENSMUST00000109912.8
ENSMUST00000128862.2 ENSMUST00000109911.8 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr8_+_110432132 | 0.05 |
ENSMUST00000212964.2
ENSMUST00000034163.9 |
Zfp821
|
zinc finger protein 821 |
| chr19_-_10807285 | 0.05 |
ENSMUST00000039043.15
|
Cd6
|
CD6 antigen |
| chr18_+_77861656 | 0.04 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr15_-_78377926 | 0.04 |
ENSMUST00000163494.3
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr11_-_102109748 | 0.04 |
ENSMUST00000131254.2
|
Hdac5
|
histone deacetylase 5 |
| chr11_+_95733109 | 0.04 |
ENSMUST00000107714.9
ENSMUST00000107711.8 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr3_+_96088467 | 0.04 |
ENSMUST00000035371.9
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr6_-_136918495 | 0.04 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr18_-_35841435 | 0.04 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr16_-_85698679 | 0.04 |
ENSMUST00000023611.7
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2) |
| chr1_-_168259839 | 0.04 |
ENSMUST00000188912.7
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr19_-_44058175 | 0.04 |
ENSMUST00000172041.8
ENSMUST00000071698.13 ENSMUST00000112028.10 |
Erlin1
|
ER lipid raft associated 1 |
| chr6_-_136918671 | 0.04 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr2_-_52448552 | 0.03 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr7_+_45084257 | 0.03 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr3_+_95190255 | 0.03 |
ENSMUST00000039537.14
ENSMUST00000107187.9 |
Mindy1
|
MINDY lysine 48 deubiquitinase 1 |
| chr16_-_92622659 | 0.03 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chrX_-_56384089 | 0.03 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr4_-_6990774 | 0.03 |
ENSMUST00000039987.4
|
Tox
|
thymocyte selection-associated high mobility group box |
| chr16_-_16681839 | 0.03 |
ENSMUST00000100136.4
|
Igll1
|
immunoglobulin lambda-like polypeptide 1 |
| chr7_-_43182504 | 0.03 |
ENSMUST00000004728.12
|
Cd33
|
CD33 antigen |
| chr8_+_110432210 | 0.03 |
ENSMUST00000212192.2
|
Zfp821
|
zinc finger protein 821 |
| chr7_+_45084300 | 0.03 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr11_-_3321307 | 0.03 |
ENSMUST00000101640.10
ENSMUST00000101642.10 |
Limk2
|
LIM motif-containing protein kinase 2 |
| chr2_-_52225146 | 0.03 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr14_+_53994813 | 0.03 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr2_+_80447389 | 0.03 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr12_-_115964081 | 0.02 |
ENSMUST00000103552.2
|
Ighv1-85
|
immunoglobulin heavy variable 1-85 |
| chr8_-_11058458 | 0.02 |
ENSMUST00000040514.8
|
Irs2
|
insulin receptor substrate 2 |
| chr2_-_119590776 | 0.02 |
ENSMUST00000082130.13
ENSMUST00000028759.13 |
Ltk
|
leukocyte tyrosine kinase |
| chr8_+_95113066 | 0.02 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chrX_-_47123719 | 0.02 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr19_-_44057800 | 0.02 |
ENSMUST00000170801.8
|
Erlin1
|
ER lipid raft associated 1 |
| chr10_+_118276949 | 0.02 |
ENSMUST00000068592.5
|
Ifng
|
interferon gamma |
| chr14_+_71011744 | 0.02 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr17_+_7592045 | 0.02 |
ENSMUST00000095726.11
ENSMUST00000128533.8 ENSMUST00000129709.8 ENSMUST00000147803.8 ENSMUST00000140192.8 ENSMUST00000138222.8 ENSMUST00000144861.2 |
Tcp10a
|
t-complex protein 10a |
| chr13_+_54937190 | 0.02 |
ENSMUST00000026993.14
ENSMUST00000131692.9 ENSMUST00000163796.8 |
Tspan17
|
tetraspanin 17 |
| chr1_-_168259264 | 0.02 |
ENSMUST00000176790.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr2_+_153780138 | 0.02 |
ENSMUST00000109757.8
ENSMUST00000154281.3 |
Bpifb4
|
BPI fold containing family B, member 4 |
| chr1_-_168259465 | 0.02 |
ENSMUST00000176540.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr11_+_6511133 | 0.02 |
ENSMUST00000160633.8
ENSMUST00000109721.3 |
Ccm2
|
cerebral cavernous malformation 2 |
| chr4_-_14621805 | 0.02 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr16_+_88828587 | 0.02 |
ENSMUST00000076906.2
|
Krtap6-1
|
keratin associated protein 6-1 |
| chr3_-_86049988 | 0.02 |
ENSMUST00000029722.7
|
Rps3a1
|
ribosomal protein S3A1 |
| chr2_+_164302863 | 0.02 |
ENSMUST00000072452.11
|
Sys1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr17_+_3447465 | 0.02 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr1_-_168259710 | 0.02 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr1_-_138102972 | 0.02 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr16_-_58539433 | 0.02 |
ENSMUST00000089318.5
|
Gpr15
|
G protein-coupled receptor 15 |
| chr10_-_120815232 | 0.02 |
ENSMUST00000119944.8
ENSMUST00000119093.2 |
Lemd3
|
LEM domain containing 3 |
| chr1_-_20854490 | 0.02 |
ENSMUST00000039046.10
|
Il17f
|
interleukin 17F |
| chr11_+_11414256 | 0.02 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr12_+_73984427 | 0.02 |
ENSMUST00000221833.2
|
Snapc1l
|
small nuclear RNA activating complex, polypeptide 1 like |
| chr8_-_96161414 | 0.02 |
ENSMUST00000211908.2
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr10_+_116111441 | 0.02 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr1_-_138103021 | 0.02 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr12_-_115276219 | 0.02 |
ENSMUST00000103529.4
|
Ighv1-58
|
immunoglobulin heavy variable 1-58 |
| chr1_+_88234454 | 0.02 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr2_-_115896279 | 0.02 |
ENSMUST00000110907.8
ENSMUST00000110908.9 |
Meis2
|
Meis homeobox 2 |
| chr5_+_139392142 | 0.02 |
ENSMUST00000052176.9
|
C130050O18Rik
|
RIKEN cDNA C130050O18 gene |
| chr19_-_10807220 | 0.02 |
ENSMUST00000174176.3
|
Cd6
|
CD6 antigen |
| chr7_-_30292351 | 0.02 |
ENSMUST00000108151.3
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr4_-_110149916 | 0.02 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr16_-_38253507 | 0.01 |
ENSMUST00000002926.8
|
Pla1a
|
phospholipase A1 member A |
| chr11_-_46280281 | 0.01 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr17_+_75485791 | 0.01 |
ENSMUST00000135447.8
ENSMUST00000112516.8 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr10_+_69761784 | 0.01 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr7_-_141014445 | 0.01 |
ENSMUST00000133021.2
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr12_+_95658987 | 0.01 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr10_-_39901249 | 0.01 |
ENSMUST00000163705.3
|
Mfsd4b1
|
major facilitator superfamily domain containing 4B1 |
| chr2_-_111320501 | 0.01 |
ENSMUST00000099616.2
|
Olfr1290
|
olfactory receptor 1290 |
| chr1_+_34199333 | 0.01 |
ENSMUST00000183302.6
ENSMUST00000185897.7 ENSMUST00000185269.7 |
Dst
|
dystonin |
| chr17_+_75485906 | 0.01 |
ENSMUST00000112514.2
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr2_-_25911544 | 0.01 |
ENSMUST00000136750.3
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr17_-_90763300 | 0.01 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr17_-_37481536 | 0.01 |
ENSMUST00000174673.3
|
Olfr753-ps1
|
olfactory receptor 753, pseudogene 1 |
| chr5_-_72893941 | 0.01 |
ENSMUST00000169534.6
|
Txk
|
TXK tyrosine kinase |
| chr6_+_41118120 | 0.01 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr1_+_152275575 | 0.01 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr4_-_14621497 | 0.01 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr2_-_86938729 | 0.01 |
ENSMUST00000099862.2
|
Olfr259
|
olfactory receptor 259 |
| chr13_-_21823691 | 0.01 |
ENSMUST00000043081.3
|
Olfr11
|
olfactory receptor 11 |
| chr10_-_114638202 | 0.01 |
ENSMUST00000239411.2
|
Trhde
|
TRH-degrading enzyme |
| chr3_+_64884839 | 0.01 |
ENSMUST00000239069.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr11_+_115197980 | 0.01 |
ENSMUST00000055490.9
|
Otop2
|
otopetrin 2 |
| chr4_+_127881786 | 0.01 |
ENSMUST00000184063.3
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr9_-_37580478 | 0.01 |
ENSMUST00000011262.4
|
Panx3
|
pannexin 3 |
| chr18_-_35782412 | 0.01 |
ENSMUST00000025211.6
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr1_-_83037090 | 0.01 |
ENSMUST00000193762.2
|
A030005K14Rik
|
RIKEN cDNA A030005K14 gene |
| chr19_-_13291647 | 0.01 |
ENSMUST00000080142.2
|
Olfr1465
|
olfactory receptor 1465 |
| chr15_-_37459570 | 0.01 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chr13_-_58423461 | 0.01 |
ENSMUST00000223811.2
|
Gkap1
|
G kinase anchoring protein 1 |
| chr14_-_124914516 | 0.01 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chr10_+_86893907 | 0.01 |
ENSMUST00000189456.7
ENSMUST00000169849.3 |
1700113H08Rik
|
RIKEN cDNA 1700113H08 gene |
| chr5_+_115568638 | 0.01 |
ENSMUST00000131079.8
|
Msi1
|
musashi RNA-binding protein 1 |
| chr6_-_83047206 | 0.01 |
ENSMUST00000174674.3
ENSMUST00000089641.6 |
Tlx2
|
T cell leukemia, homeobox 2 |
| chr7_+_51537645 | 0.01 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr15_-_82648376 | 0.01 |
ENSMUST00000055721.6
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr5_+_37025810 | 0.01 |
ENSMUST00000031003.11
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chrX_+_158480304 | 0.01 |
ENSMUST00000123433.8
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr2_+_32519479 | 0.01 |
ENSMUST00000068271.5
|
Ak1
|
adenylate kinase 1 |
| chr13_-_21859150 | 0.01 |
ENSMUST00000079135.6
|
Olfr1360
|
olfactory receptor 1360 |
| chr2_+_21372338 | 0.01 |
ENSMUST00000055946.8
|
Gpr158
|
G protein-coupled receptor 158 |
| chr4_-_137137088 | 0.01 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr13_+_21316868 | 0.01 |
ENSMUST00000096006.3
|
Olfr263
|
olfactory receptor 263 |
| chr3_-_33136153 | 0.01 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr7_+_119289249 | 0.01 |
ENSMUST00000047045.10
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr5_+_37025926 | 0.01 |
ENSMUST00000201156.2
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr2_+_20727274 | 0.01 |
ENSMUST00000114607.8
|
Etl4
|
enhancer trap locus 4 |
| chr6_+_126830050 | 0.01 |
ENSMUST00000095440.9
|
Akap3
|
A kinase (PRKA) anchor protein 3 |
| chr7_-_141794815 | 0.01 |
ENSMUST00000211591.2
|
Gm40460
|
predicted gene, 40460 |
| chr19_+_4264292 | 0.01 |
ENSMUST00000046506.7
|
Clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr11_-_100098333 | 0.01 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr18_-_60781365 | 0.01 |
ENSMUST00000143275.3
|
Synpo
|
synaptopodin |
| chr10_+_127575407 | 0.01 |
ENSMUST00000054287.9
|
Zbtb39
|
zinc finger and BTB domain containing 39 |
| chr17_+_14087827 | 0.01 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr2_-_89774457 | 0.01 |
ENSMUST00000090695.3
|
Olfr1259
|
olfactory receptor 1259 |
| chr6_+_126830102 | 0.01 |
ENSMUST00000202878.4
ENSMUST00000202574.2 |
Akap3
|
A kinase (PRKA) anchor protein 3 |
| chr15_-_37734579 | 0.01 |
ENSMUST00000145909.9
ENSMUST00000153775.9 |
Gm49397
Ncald
|
predicted gene, 49397 neurocalcin delta |
| chr1_-_161616031 | 0.01 |
ENSMUST00000000834.4
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr2_+_111610658 | 0.01 |
ENSMUST00000054004.2
|
Olfr1302
|
olfactory receptor 1302 |
| chr16_+_25620652 | 0.01 |
ENSMUST00000115304.8
ENSMUST00000115305.2 ENSMUST00000040231.13 ENSMUST00000115306.8 |
Trp63
|
transformation related protein 63 |
| chr15_+_9436114 | 0.01 |
ENSMUST00000042360.5
ENSMUST00000226688.2 |
Capsl
|
calcyphosine-like |
| chr1_+_82891043 | 0.01 |
ENSMUST00000220768.2
|
A030005L19Rik
|
RIKEN cDNA A030005L19 gene |
| chr2_+_153760311 | 0.01 |
ENSMUST00000109760.2
|
Bpifb3
|
BPI fold containing family B, member 3 |
| chr9_+_108356935 | 0.01 |
ENSMUST00000194147.2
ENSMUST00000065014.10 ENSMUST00000195483.6 ENSMUST00000195058.2 |
Lamb2
|
laminin, beta 2 |
| chr3_-_151960948 | 0.01 |
ENSMUST00000199423.5
ENSMUST00000198460.5 |
Nexn
|
nexilin |
| chr4_-_118639783 | 0.01 |
ENSMUST00000077247.3
|
Olfr1337
|
olfactory receptor 1337 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.3 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.4 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.0 | 0.2 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.0 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |