Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Runx2_Bcl11a
Z-value: 0.47
Transcription factors associated with Runx2_Bcl11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
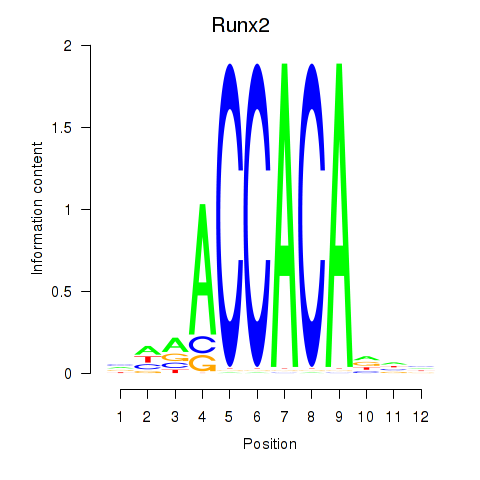
Runx2
|
ENSMUSG00000039153.18 | runt related transcription factor 2 |
|
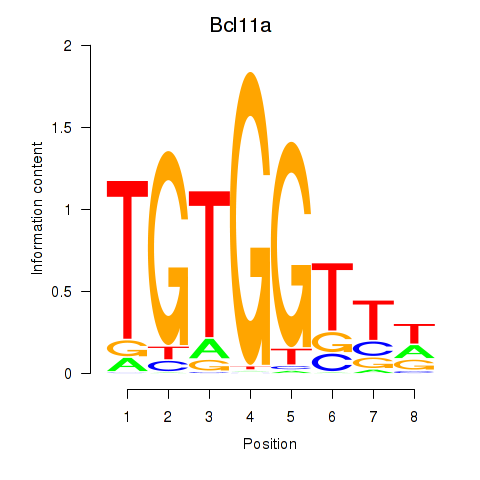
Bcl11a
|
ENSMUSG00000000861.16 | B cell CLL/lymphoma 11A (zinc finger protein) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx2 | mm39_v1_chr17_-_45046499_45046597 | 0.70 | 1.9e-01 | Click! |
| Bcl11a | mm39_v1_chr11_+_24028022_24028076 | 0.53 | 3.6e-01 | Click! |
Activity profile of Runx2_Bcl11a motif
Sorted Z-values of Runx2_Bcl11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_162687488 | 0.43 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr17_-_45125468 | 0.39 |
ENSMUST00000159943.8
ENSMUST00000160673.8 |
Runx2
|
runt related transcription factor 2 |
| chr2_-_121211410 | 0.36 |
ENSMUST00000038389.15
|
Strc
|
stereocilin |
| chr16_+_32090286 | 0.35 |
ENSMUST00000093183.5
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr18_+_39439778 | 0.35 |
ENSMUST00000235660.2
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr1_-_173569301 | 0.33 |
ENSMUST00000042610.15
ENSMUST00000127730.2 |
Ifi207
|
interferon activated gene 207 |
| chr16_-_92622972 | 0.32 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr15_+_6599001 | 0.31 |
ENSMUST00000227175.2
|
Fyb
|
FYN binding protein |
| chr9_-_39863342 | 0.31 |
ENSMUST00000216647.2
|
Olfr975
|
olfactory receptor 975 |
| chr14_-_51045182 | 0.30 |
ENSMUST00000227614.2
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr3_+_103767581 | 0.30 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr13_-_23882437 | 0.29 |
ENSMUST00000102967.3
|
H4c3
|
H4 clustered histone 3 |
| chr6_+_136931519 | 0.29 |
ENSMUST00000137768.2
|
Pde6h
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr11_+_105866030 | 0.28 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr15_-_53765869 | 0.28 |
ENSMUST00000078673.14
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr11_-_79414542 | 0.27 |
ENSMUST00000179322.2
|
Evi2b
|
ecotropic viral integration site 2b |
| chr7_-_5548308 | 0.27 |
ENSMUST00000236262.2
|
Vmn1r60
|
vomeronasal 1 receptor 60 |
| chr13_+_21938258 | 0.27 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr4_+_132903646 | 0.24 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr7_-_3828640 | 0.24 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chr7_+_101011204 | 0.24 |
ENSMUST00000133423.2
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr7_-_126275529 | 0.24 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr15_+_6552270 | 0.23 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr7_-_140596811 | 0.23 |
ENSMUST00000081924.5
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr7_-_141241632 | 0.22 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr2_-_26184450 | 0.22 |
ENSMUST00000217256.2
ENSMUST00000227200.2 |
Ccdc187
|
coiled-coil domain containing 187 |
| chr2_-_88947627 | 0.22 |
ENSMUST00000217635.2
ENSMUST00000143255.3 ENSMUST00000213404.2 |
Olfr1221
|
olfactory receptor 1221 |
| chr17_+_33857030 | 0.22 |
ENSMUST00000052079.8
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1 |
| chr15_+_98065039 | 0.21 |
ENSMUST00000031914.6
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr15_+_76238632 | 0.20 |
ENSMUST00000208833.3
|
Gm35339
|
predicted gene, 35339 |
| chr8_+_84852609 | 0.20 |
ENSMUST00000093380.5
|
Podnl1
|
podocan-like 1 |
| chr6_+_113366207 | 0.20 |
ENSMUST00000204026.3
|
Ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chrX_-_20157966 | 0.20 |
ENSMUST00000115393.3
ENSMUST00000072451.11 |
Slc9a7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7 |
| chr8_-_3744167 | 0.20 |
ENSMUST00000005678.6
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide |
| chr5_-_138153956 | 0.19 |
ENSMUST00000132318.2
ENSMUST00000049393.15 |
Zfp113
|
zinc finger protein 113 |
| chr7_+_5023375 | 0.19 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865 |
| chr7_+_30159137 | 0.18 |
ENSMUST00000006825.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr13_+_22508759 | 0.18 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr1_-_72914036 | 0.17 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr2_-_34990689 | 0.17 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr13_-_23805077 | 0.17 |
ENSMUST00000091704.7
ENSMUST00000051091.5 |
H2bc6
|
H2B clustered histone 6 |
| chr14_+_50656083 | 0.17 |
ENSMUST00000216949.2
|
Olfr739
|
olfactory receptor 739 |
| chr5_+_145063568 | 0.17 |
ENSMUST00000138922.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr7_-_3848050 | 0.17 |
ENSMUST00000108615.10
ENSMUST00000119469.2 |
Pira2
|
paired-Ig-like receptor A2 |
| chr7_-_43182595 | 0.17 |
ENSMUST00000205503.2
|
Cd33
|
CD33 antigen |
| chr17_+_35235552 | 0.16 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr16_+_35977170 | 0.16 |
ENSMUST00000079184.6
|
Stfa2l1
|
stefin A2 like 1 |
| chr12_+_33365371 | 0.16 |
ENSMUST00000154742.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr19_+_8828132 | 0.16 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr19_+_8975249 | 0.15 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr5_-_115479203 | 0.15 |
ENSMUST00000139167.3
|
Gatc
|
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr16_-_92622659 | 0.15 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr7_-_24831892 | 0.15 |
ENSMUST00000108418.11
ENSMUST00000175774.9 ENSMUST00000108415.10 ENSMUST00000098679.10 ENSMUST00000108417.10 ENSMUST00000108416.10 ENSMUST00000108413.8 ENSMUST00000176408.8 |
Pou2f2
|
POU domain, class 2, transcription factor 2 |
| chr3_-_15397325 | 0.15 |
ENSMUST00000108361.2
|
Gm9733
|
predicted gene 9733 |
| chr10_+_127612243 | 0.15 |
ENSMUST00000136223.2
ENSMUST00000052652.7 |
Rdh9
|
retinol dehydrogenase 9 |
| chr5_-_113957362 | 0.14 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr19_-_5323092 | 0.14 |
ENSMUST00000237463.2
ENSMUST00000025786.9 |
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chrX_-_139443926 | 0.14 |
ENSMUST00000055738.12
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr11_-_54751738 | 0.14 |
ENSMUST00000144164.9
|
Lyrm7
|
LYR motif containing 7 |
| chr7_-_103113358 | 0.14 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607 |
| chr7_-_3901119 | 0.14 |
ENSMUST00000070639.8
|
Gm14548
|
predicted gene 14548 |
| chr16_-_18880821 | 0.14 |
ENSMUST00000200568.2
|
Iglc1
|
immunoglobulin lambda constant 1 |
| chr17_+_24022153 | 0.14 |
ENSMUST00000190686.7
ENSMUST00000088621.11 ENSMUST00000233636.2 |
Srrm2
|
serine/arginine repetitive matrix 2 |
| chrX_+_158410528 | 0.14 |
ENSMUST00000073094.10
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chrX_+_105070907 | 0.13 |
ENSMUST00000055941.7
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr9_-_51076724 | 0.13 |
ENSMUST00000210433.2
|
Gm32742
|
predicted gene, 32742 |
| chr11_+_117545037 | 0.13 |
ENSMUST00000026658.13
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr10_-_80382611 | 0.13 |
ENSMUST00000183233.9
ENSMUST00000182604.8 |
Rexo1
|
REX1, RNA exonuclease 1 |
| chr5_+_105667254 | 0.13 |
ENSMUST00000067924.13
ENSMUST00000150981.2 |
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr8_+_71207326 | 0.13 |
ENSMUST00000110093.9
ENSMUST00000143118.3 ENSMUST00000034301.12 ENSMUST00000110090.8 |
Rab3a
|
RAB3A, member RAS oncogene family |
| chr2_-_73316809 | 0.13 |
ENSMUST00000141264.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr11_+_101932328 | 0.13 |
ENSMUST00000123895.8
ENSMUST00000017453.12 ENSMUST00000107163.9 ENSMUST00000107164.3 |
Cd300lg
|
CD300 molecule like family member G |
| chr3_-_129834788 | 0.12 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr12_+_116041340 | 0.12 |
ENSMUST00000011315.10
|
Vipr2
|
vasoactive intestinal peptide receptor 2 |
| chr7_-_3723381 | 0.12 |
ENSMUST00000078451.7
|
Pirb
|
paired Ig-like receptor B |
| chr10_-_12689345 | 0.12 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr14_-_32907446 | 0.12 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr4_+_45890303 | 0.12 |
ENSMUST00000178561.8
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr17_+_12338161 | 0.12 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr1_+_107456731 | 0.12 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr11_+_69657275 | 0.12 |
ENSMUST00000132528.8
ENSMUST00000153943.2 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr18_+_62681982 | 0.12 |
ENSMUST00000055725.12
ENSMUST00000162365.8 |
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr3_-_88280047 | 0.12 |
ENSMUST00000107543.8
ENSMUST00000107542.2 |
Bglap3
|
bone gamma-carboxyglutamate protein 3 |
| chr10_+_127595590 | 0.12 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr17_+_15262510 | 0.11 |
ENSMUST00000226561.2
|
Ermard
|
ER membrane associated RNA degradation |
| chr1_-_160134873 | 0.11 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chrX_+_98046829 | 0.11 |
ENSMUST00000149999.8
|
Stard8
|
START domain containing 8 |
| chr4_-_98712077 | 0.11 |
ENSMUST00000097964.3
|
I0C0044D17Rik
|
RIKEN cDNA I0C0044D17 gene |
| chr8_-_123885007 | 0.11 |
ENSMUST00000000755.15
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr5_-_91550853 | 0.11 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr10_+_127226180 | 0.11 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chr5_+_64969679 | 0.11 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr7_+_100355910 | 0.11 |
ENSMUST00000207875.2
ENSMUST00000208013.2 |
Fam168a
|
family with sequence similarity 168, member A |
| chr7_-_43182504 | 0.11 |
ENSMUST00000004728.12
|
Cd33
|
CD33 antigen |
| chr7_+_26895206 | 0.11 |
ENSMUST00000179391.8
ENSMUST00000108379.8 |
BC024978
|
cDNA sequence BC024978 |
| chr11_+_75422516 | 0.11 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr6_+_123100382 | 0.11 |
ENSMUST00000032248.8
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr17_+_35286293 | 0.10 |
ENSMUST00000173478.2
ENSMUST00000174876.2 |
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr4_+_134847949 | 0.10 |
ENSMUST00000056977.14
|
Runx3
|
runt related transcription factor 3 |
| chr13_+_54519161 | 0.10 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chr2_-_28589675 | 0.10 |
ENSMUST00000124840.2
|
Spaca9
|
sperm acrosome associated 9 |
| chr2_-_28589641 | 0.10 |
ENSMUST00000102877.8
|
Spaca9
|
sperm acrosome associated 9 |
| chrX_-_73009933 | 0.10 |
ENSMUST00000114372.3
ENSMUST00000033761.13 |
Hcfc1
|
host cell factor C1 |
| chr9_+_108880158 | 0.10 |
ENSMUST00000198708.5
|
Shisa5
|
shisa family member 5 |
| chr9_+_119978773 | 0.10 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr7_-_127423641 | 0.10 |
ENSMUST00000106267.5
|
Stx1b
|
syntaxin 1B |
| chr2_+_88217406 | 0.10 |
ENSMUST00000214040.3
|
Olfr1178
|
olfactory receptor 1178 |
| chr12_+_108601963 | 0.10 |
ENSMUST00000223109.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr6_+_116627567 | 0.10 |
ENSMUST00000067354.10
ENSMUST00000178241.4 |
Depp1
|
DEPP1 autophagy regulator |
| chr8_+_3705760 | 0.10 |
ENSMUST00000169234.3
ENSMUST00000012849.15 |
Retn
|
resistin |
| chr17_-_38370970 | 0.10 |
ENSMUST00000216476.2
|
Olfr129
|
olfactory receptor 129 |
| chr1_+_90926443 | 0.10 |
ENSMUST00000189505.7
ENSMUST00000185531.7 ENSMUST00000068116.13 |
Lrrfip1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chr7_+_125307116 | 0.10 |
ENSMUST00000148701.4
|
Katnip
|
katanin interacting protein |
| chr9_+_39932760 | 0.10 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr1_+_173093568 | 0.10 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr16_-_36228798 | 0.10 |
ENSMUST00000023619.8
|
Stfa2
|
stefin A2 |
| chr17_+_48623157 | 0.10 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr7_-_126183716 | 0.10 |
ENSMUST00000150311.8
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr11_+_117545618 | 0.10 |
ENSMUST00000106344.8
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr7_+_127475968 | 0.10 |
ENSMUST00000131000.2
|
Zfp646
|
zinc finger protein 646 |
| chr7_-_46445305 | 0.09 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr1_-_95595245 | 0.09 |
ENSMUST00000189556.2
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr10_-_30076488 | 0.09 |
ENSMUST00000216853.2
|
Cenpw
|
centromere protein W |
| chr9_+_102988940 | 0.09 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr6_+_123100272 | 0.09 |
ENSMUST00000041779.13
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr10_-_85847697 | 0.09 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr13_-_54835460 | 0.09 |
ENSMUST00000129881.8
|
Rnf44
|
ring finger protein 44 |
| chr11_-_62349334 | 0.09 |
ENSMUST00000141447.2
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr1_-_170133901 | 0.09 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694 |
| chr11_+_49176109 | 0.09 |
ENSMUST00000217275.2
ENSMUST00000214598.2 ENSMUST00000215861.2 ENSMUST00000214170.2 |
Olfr1392
|
olfactory receptor 1392 |
| chr9_-_110474398 | 0.09 |
ENSMUST00000149089.2
|
Nbeal2
|
neurobeachin-like 2 |
| chr7_-_14297431 | 0.09 |
ENSMUST00000227855.2
ENSMUST00000185220.3 ENSMUST00000226802.2 ENSMUST00000227692.2 ENSMUST00000227788.2 ENSMUST00000227566.2 ENSMUST00000226510.2 ENSMUST00000226264.2 |
Vmn1r90
|
vomeronasal 1 receptor 90 |
| chr7_-_43906802 | 0.09 |
ENSMUST00000107945.8
ENSMUST00000118216.8 |
Acp4
|
acid phosphatase 4 |
| chr7_-_24705320 | 0.09 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr7_+_100355798 | 0.09 |
ENSMUST00000107042.9
ENSMUST00000207564.2 ENSMUST00000049053.9 |
Fam168a
|
family with sequence similarity 168, member A |
| chr9_+_108880221 | 0.09 |
ENSMUST00000200629.5
ENSMUST00000200515.5 ENSMUST00000197689.5 ENSMUST00000196954.5 ENSMUST00000198376.5 ENSMUST00000197483.5 ENSMUST00000198295.5 |
Shisa5
|
shisa family member 5 |
| chr17_-_45125537 | 0.09 |
ENSMUST00000113571.10
|
Runx2
|
runt related transcription factor 2 |
| chr11_-_106469938 | 0.09 |
ENSMUST00000103070.3
|
Tex2
|
testis expressed gene 2 |
| chr1_+_16758629 | 0.09 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr3_-_59009231 | 0.09 |
ENSMUST00000085040.5
|
Gpr171
|
G protein-coupled receptor 171 |
| chrX_+_16485937 | 0.09 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr2_-_73316053 | 0.09 |
ENSMUST00000102680.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr7_+_5023552 | 0.08 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
| chr12_-_110649040 | 0.08 |
ENSMUST00000222915.2
ENSMUST00000070659.7 |
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr16_-_94023976 | 0.08 |
ENSMUST00000227698.2
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase) |
| chr11_+_115921129 | 0.08 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr11_+_75422925 | 0.08 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr7_+_127845984 | 0.08 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr2_+_172235702 | 0.08 |
ENSMUST00000099061.9
ENSMUST00000103073.9 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr1_-_84262144 | 0.08 |
ENSMUST00000176720.2
|
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr1_+_170104889 | 0.08 |
ENSMUST00000179976.3
|
Sh2d1b1
|
SH2 domain containing 1B1 |
| chr10_+_3822667 | 0.08 |
ENSMUST00000136671.8
ENSMUST00000042438.13 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chrM_+_14138 | 0.08 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr9_-_39486831 | 0.08 |
ENSMUST00000216298.2
ENSMUST00000215194.2 |
Olfr959
|
olfactory receptor 959 |
| chr3_+_96127174 | 0.08 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21 |
| chr17_+_37269468 | 0.08 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr14_-_122153185 | 0.08 |
ENSMUST00000055475.9
|
Gpr18
|
G protein-coupled receptor 18 |
| chr5_+_114427227 | 0.08 |
ENSMUST00000169347.6
|
Myo1h
|
myosin 1H |
| chr15_-_102630589 | 0.08 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr6_+_126916919 | 0.08 |
ENSMUST00000032497.7
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr11_+_96740689 | 0.08 |
ENSMUST00000018816.14
|
Copz2
|
coatomer protein complex, subunit zeta 2 |
| chrX_-_72913410 | 0.08 |
ENSMUST00000066576.12
ENSMUST00000114430.8 |
L1cam
|
L1 cell adhesion molecule |
| chr9_-_110818679 | 0.08 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr7_+_3620356 | 0.08 |
ENSMUST00000076657.11
ENSMUST00000108644.8 |
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr6_-_67468831 | 0.08 |
ENSMUST00000118364.2
|
Il23r
|
interleukin 23 receptor |
| chrM_-_14061 | 0.08 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr1_-_192880260 | 0.08 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr7_+_119495058 | 0.08 |
ENSMUST00000106518.9
ENSMUST00000207270.2 ENSMUST00000208424.2 ENSMUST00000208202.2 ENSMUST00000054440.11 |
Lyrm1
|
LYR motif containing 1 |
| chr4_-_155953849 | 0.08 |
ENSMUST00000030950.8
|
Cptp
|
ceramide-1-phosphate transfer protein |
| chr7_+_125307060 | 0.08 |
ENSMUST00000124223.8
ENSMUST00000069660.13 |
Katnip
|
katanin interacting protein |
| chr7_+_107585900 | 0.08 |
ENSMUST00000214677.2
|
Olfr477
|
olfactory receptor 477 |
| chr9_+_121471782 | 0.07 |
ENSMUST00000035115.5
|
Vipr1
|
vasoactive intestinal peptide receptor 1 |
| chr7_+_24048613 | 0.07 |
ENSMUST00000032683.6
|
Lypd5
|
Ly6/Plaur domain containing 5 |
| chr14_-_20027279 | 0.07 |
ENSMUST00000160013.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr2_+_11647610 | 0.07 |
ENSMUST00000028111.6
|
Il2ra
|
interleukin 2 receptor, alpha chain |
| chr10_+_51356728 | 0.07 |
ENSMUST00000102894.6
ENSMUST00000219661.2 ENSMUST00000219696.2 ENSMUST00000217706.2 |
Lilr4b
Gm49339
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4B predicted gene, 49339 |
| chr5_-_140687995 | 0.07 |
ENSMUST00000135028.5
ENSMUST00000077890.12 ENSMUST00000041783.14 ENSMUST00000142081.6 |
Iqce
|
IQ motif containing E |
| chr17_-_48474356 | 0.07 |
ENSMUST00000027764.10
ENSMUST00000053612.14 |
A530064D06Rik
|
RIKEN cDNA A530064D06 gene |
| chr5_+_122781941 | 0.07 |
ENSMUST00000100737.10
ENSMUST00000121489.8 ENSMUST00000031425.15 ENSMUST00000086247.6 |
P2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr15_-_68130636 | 0.07 |
ENSMUST00000162173.8
ENSMUST00000160248.8 ENSMUST00000162054.9 |
Zfat
|
zinc finger and AT hook domain containing |
| chr5_+_150119860 | 0.07 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr17_-_36290129 | 0.07 |
ENSMUST00000165613.9
ENSMUST00000173872.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr18_-_62313019 | 0.07 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr15_+_3300249 | 0.07 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr9_-_53617508 | 0.07 |
ENSMUST00000068449.4
|
Rab39
|
RAB39, member RAS oncogene family |
| chr13_+_74086241 | 0.07 |
ENSMUST00000222749.2
|
Brd9
|
bromodomain containing 9 |
| chr17_+_35454833 | 0.07 |
ENSMUST00000118384.8
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr17_+_35455532 | 0.07 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr4_-_127864744 | 0.07 |
ENSMUST00000030614.3
|
CK137956
|
cDNA sequence CK137956 |
| chr10_-_12744025 | 0.07 |
ENSMUST00000219660.2
|
Utrn
|
utrophin |
| chr4_-_114991478 | 0.07 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr7_+_79939747 | 0.07 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr16_+_91343936 | 0.07 |
ENSMUST00000023687.9
|
Ifngr2
|
interferon gamma receptor 2 |
| chrX_-_9335525 | 0.07 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr1_-_171061902 | 0.07 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr1_+_153750081 | 0.07 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chr7_-_140676623 | 0.07 |
ENSMUST00000209352.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr17_+_43700327 | 0.07 |
ENSMUST00000113599.2
ENSMUST00000224278.2 ENSMUST00000225466.2 |
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chrX_+_162692126 | 0.07 |
ENSMUST00000033734.14
ENSMUST00000112294.9 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_+_62349238 | 0.07 |
ENSMUST00000014389.6
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx2_Bcl11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.1 | 0.3 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.3 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 | 0.2 | GO:1904959 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.3 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 0.7 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.1 | GO:1904048 | negative regulation of synaptic vesicle recycling(GO:1903422) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:0072249 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.0 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.0 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0090096 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.0 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.0 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.1 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.2 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.0 | 0.1 | GO:0004080 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.0 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.0 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.0 | 0.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |