Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
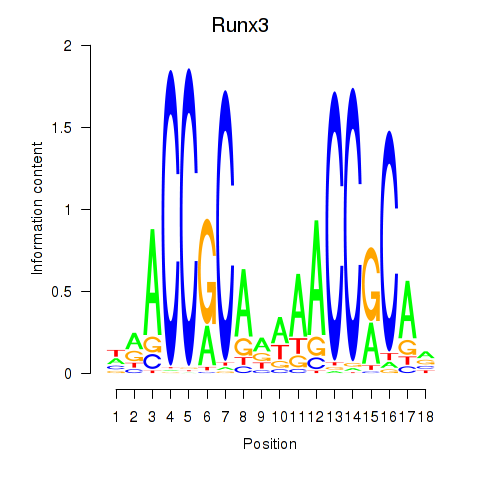
Results for Runx3
Z-value: 1.76
Transcription factors associated with Runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx3
|
ENSMUSG00000070691.11 | runt related transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx3 | mm39_v1_chr4_+_134879807_134879865 | 0.76 | 1.3e-01 | Click! |
Activity profile of Runx3 motif
Sorted Z-values of Runx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_23882437 | 5.18 |
ENSMUST00000102967.3
|
H4c3
|
H4 clustered histone 3 |
| chr13_-_23735822 | 4.62 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr13_-_22016364 | 3.13 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr13_+_21919225 | 3.10 |
ENSMUST00000087714.6
|
H4c11
|
H4 clustered histone 11 |
| chr13_-_21934675 | 3.05 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr6_-_136781406 | 2.79 |
ENSMUST00000179285.3
|
H4f16
|
H4 histone 16 |
| chr13_-_22225527 | 1.39 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9 |
| chr14_-_98406977 | 1.00 |
ENSMUST00000071533.13
ENSMUST00000069334.8 |
Dach1
|
dachshund family transcription factor 1 |
| chrX_-_97934387 | 0.81 |
ENSMUST00000113826.8
ENSMUST00000033560.9 ENSMUST00000142267.2 |
Ophn1
|
oligophrenin 1 |
| chr5_-_36987917 | 0.80 |
ENSMUST00000031002.10
|
Man2b2
|
mannosidase 2, alpha B2 |
| chr12_+_44315927 | 0.78 |
ENSMUST00000043082.16
ENSMUST00000218954.2 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr9_+_106306736 | 0.73 |
ENSMUST00000098994.7
ENSMUST00000059802.7 ENSMUST00000213448.2 ENSMUST00000217081.2 |
Rpl29
|
ribosomal protein L29 |
| chr2_+_37080286 | 0.70 |
ENSMUST00000218602.2
|
Olfr365
|
olfactory receptor 365 |
| chr9_+_106306598 | 0.69 |
ENSMUST00000150576.8
|
Rpl29
|
ribosomal protein L29 |
| chr19_-_20368029 | 0.57 |
ENSMUST00000235280.2
|
Anxa1
|
annexin A1 |
| chr13_-_115238427 | 0.56 |
ENSMUST00000224997.2
ENSMUST00000061673.9 |
Gm49395
Itga1
|
predicted gene, 49395 integrin alpha 1 |
| chrX_+_162691978 | 0.52 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr16_-_10131804 | 0.43 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2 |
| chr13_+_38009981 | 0.42 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr10_+_42378193 | 0.40 |
ENSMUST00000105499.2
|
Snx3
|
sorting nexin 3 |
| chr9_+_109760931 | 0.39 |
ENSMUST00000165876.8
|
Map4
|
microtubule-associated protein 4 |
| chr15_-_78739717 | 0.39 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr9_+_109760528 | 0.38 |
ENSMUST00000035055.15
|
Map4
|
microtubule-associated protein 4 |
| chr14_-_72946972 | 0.38 |
ENSMUST00000162478.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr5_+_135382149 | 0.38 |
ENSMUST00000111180.9
ENSMUST00000065785.4 |
Trim50
|
tripartite motif-containing 50 |
| chr1_-_24139263 | 0.35 |
ENSMUST00000187369.7
ENSMUST00000187752.7 ENSMUST00000186999.7 |
Fam135a
|
family with sequence similarity 135, member A |
| chr2_+_145627900 | 0.34 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr4_+_19575128 | 0.33 |
ENSMUST00000108253.8
ENSMUST00000029888.4 |
Rmdn1
|
regulator of microtubule dynamics 1 |
| chr7_+_126690525 | 0.29 |
ENSMUST00000056288.7
ENSMUST00000206102.2 |
AI467606
|
expressed sequence AI467606 |
| chr17_+_28547548 | 0.29 |
ENSMUST00000233895.2
ENSMUST00000232867.2 |
Rpl10a
|
ribosomal protein L10A |
| chr13_-_51888737 | 0.24 |
ENSMUST00000110039.2
|
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr9_+_109760856 | 0.21 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr18_-_47501513 | 0.19 |
ENSMUST00000076043.13
ENSMUST00000135790.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr1_+_40505136 | 0.18 |
ENSMUST00000087983.8
ENSMUST00000195684.6 ENSMUST00000108044.4 |
Il18r1
|
interleukin 18 receptor 1 |
| chr3_-_7678785 | 0.16 |
ENSMUST00000194279.6
|
Il7
|
interleukin 7 |
| chr16_+_4825216 | 0.16 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr1_-_24139387 | 0.15 |
ENSMUST00000027337.15
|
Fam135a
|
family with sequence similarity 135, member A |
| chr9_+_110361561 | 0.15 |
ENSMUST00000153838.8
|
Setd2
|
SET domain containing 2 |
| chr12_-_98225676 | 0.15 |
ENSMUST00000021390.9
|
Galc
|
galactosylceramidase |
| chr17_+_27775471 | 0.14 |
ENSMUST00000118599.9
ENSMUST00000232265.2 ENSMUST00000232013.2 ENSMUST00000114888.11 ENSMUST00000231874.2 ENSMUST00000119486.9 ENSMUST00000231825.2 ENSMUST00000231866.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr17_+_28547445 | 0.14 |
ENSMUST00000042334.16
|
Rpl10a
|
ribosomal protein L10A |
| chr7_+_28508220 | 0.11 |
ENSMUST00000172529.8
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr11_-_5828251 | 0.11 |
ENSMUST00000102922.10
|
Pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr10_-_80837173 | 0.10 |
ENSMUST00000099462.8
ENSMUST00000118233.8 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr2_-_28356294 | 0.08 |
ENSMUST00000127683.2
ENSMUST00000086370.11 |
1700007K13Rik
|
RIKEN cDNA 1700007K13 gene |
| chr17_-_45744637 | 0.08 |
ENSMUST00000024727.10
|
Cdc5l
|
cell division cycle 5-like (S. pombe) |
| chr13_+_14238361 | 0.07 |
ENSMUST00000129488.8
ENSMUST00000110536.8 ENSMUST00000110534.8 ENSMUST00000039538.15 ENSMUST00000110533.2 |
Arid4b
|
AT rich interactive domain 4B (RBP1-like) |
| chr15_-_95553841 | 0.07 |
ENSMUST00000054244.7
|
Dbx2
|
developing brain homeobox 2 |
| chr5_-_21629661 | 0.06 |
ENSMUST00000115245.8
ENSMUST00000030552.7 |
Ccdc146
|
coiled-coil domain containing 146 |
| chr6_+_115908709 | 0.06 |
ENSMUST00000032471.9
|
Rho
|
rhodopsin |
| chr1_-_154602102 | 0.06 |
ENSMUST00000187541.7
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr13_+_46822992 | 0.06 |
ENSMUST00000099547.4
|
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr1_+_82938007 | 0.06 |
ENSMUST00000223536.2
|
Gm47955
|
predicted gene, 47955 |
| chr7_-_141710850 | 0.06 |
ENSMUST00000209599.2
|
Gm29735
|
predicted gene, 29735 |
| chr10_-_128133953 | 0.05 |
ENSMUST00000026449.3
|
Il23a
|
interleukin 23, alpha subunit p19 |
| chr2_+_155359868 | 0.05 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr13_+_38009951 | 0.05 |
ENSMUST00000138043.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr6_+_21949570 | 0.05 |
ENSMUST00000031680.10
ENSMUST00000115389.8 ENSMUST00000151473.8 |
Ing3
|
inhibitor of growth family, member 3 |
| chr17_-_23818458 | 0.04 |
ENSMUST00000057029.5
|
Zfp13
|
zinc finger protein 13 |
| chr9_-_58065800 | 0.04 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr2_-_71377088 | 0.04 |
ENSMUST00000024159.8
|
Dlx2
|
distal-less homeobox 2 |
| chr6_-_135231168 | 0.04 |
ENSMUST00000111909.8
|
Gsg1
|
germ cell associated 1 |
| chr10_+_79590910 | 0.03 |
ENSMUST00000219981.2
ENSMUST00000219228.2 ENSMUST00000020577.4 |
Fgf22
|
fibroblast growth factor 22 |
| chr7_-_44624165 | 0.03 |
ENSMUST00000212836.2
ENSMUST00000212255.2 ENSMUST00000063761.8 |
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chrX_-_146337046 | 0.03 |
ENSMUST00000112819.9
ENSMUST00000136789.8 |
Lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr7_+_141755098 | 0.03 |
ENSMUST00000187512.2
ENSMUST00000084414.6 |
Krtap5-3
|
keratin associated protein 5-3 |
| chr8_+_27937691 | 0.02 |
ENSMUST00000081321.5
|
Poteg
|
POTE ankyrin domain family, member G |
| chr2_+_85822163 | 0.02 |
ENSMUST00000050942.3
|
Olfr1031
|
olfactory receptor 1031 |
| chr7_-_141698342 | 0.02 |
ENSMUST00000210537.2
|
Gm45337
|
predicted gene 45337 |
| chr12_+_103277234 | 0.02 |
ENSMUST00000191218.7
|
Fam181a
|
family with sequence similarity 181, member A |
| chr11_-_32217547 | 0.02 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr9_+_121548469 | 0.02 |
ENSMUST00000182225.8
|
Nktr
|
natural killer tumor recognition sequence |
| chr4_-_133225849 | 0.02 |
ENSMUST00000125541.2
|
Trnp1
|
TMF1-regulated nuclear protein 1 |
| chr18_-_50701793 | 0.02 |
ENSMUST00000056460.4
|
Pudp
|
pseudouridine 5'-phosphatase |
| chr1_-_82921109 | 0.02 |
ENSMUST00000222600.2
|
A030003K21Rik
|
RIKEN cDNA A030003K21 gene |
| chr3_-_7678796 | 0.02 |
ENSMUST00000192202.6
|
Il7
|
interleukin 7 |
| chr4_-_70328659 | 0.01 |
ENSMUST00000144099.8
|
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chr15_+_98953776 | 0.01 |
ENSMUST00000229268.2
|
Prph
|
peripherin |
| chr1_+_82902636 | 0.01 |
ENSMUST00000223400.2
|
A030014E15Rik
|
RIKEN cDNA A030014E15 gene |
| chr4_+_134070492 | 0.01 |
ENSMUST00000105873.8
ENSMUST00000105874.9 |
Slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr11_-_100098333 | 0.01 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr16_-_20972750 | 0.01 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr2_+_155360015 | 0.01 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr1_+_74317709 | 0.01 |
ENSMUST00000077985.4
|
Gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr9_+_58395850 | 0.01 |
ENSMUST00000085658.5
|
Insyn1
|
inhibitory synaptic factor 1 |
| chr8_-_55177510 | 0.00 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chrY_-_6917117 | 0.00 |
ENSMUST00000178016.2
|
Gm20830
|
predicted gene, 20830 |
| chr2_+_157756535 | 0.00 |
ENSMUST00000109523.2
|
Vstm2l
|
V-set and transmembrane domain containing 2-like |
| chr7_-_141794815 | 0.00 |
ENSMUST00000211591.2
|
Gm40460
|
predicted gene, 40460 |
| chr11_-_57722830 | 0.00 |
ENSMUST00000036917.3
|
Hand1
|
heart and neural crest derivatives expressed 1 |
| chr7_-_25516041 | 0.00 |
ENSMUST00000043314.10
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr7_+_106507208 | 0.00 |
ENSMUST00000215949.2
ENSMUST00000207492.3 |
Olfr1532-ps1
|
olfactory receptor 1532, pseudogene 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.2 | 1.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.6 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.4 | GO:2001212 | caveola assembly(GO:0070836) regulation of vasculogenesis(GO:2001212) |
| 0.0 | 0.0 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.6 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:2000328 | positive regulation of memory T cell differentiation(GO:0043382) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 1.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.3 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.2 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 1.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |