Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Rxra
Z-value: 0.31
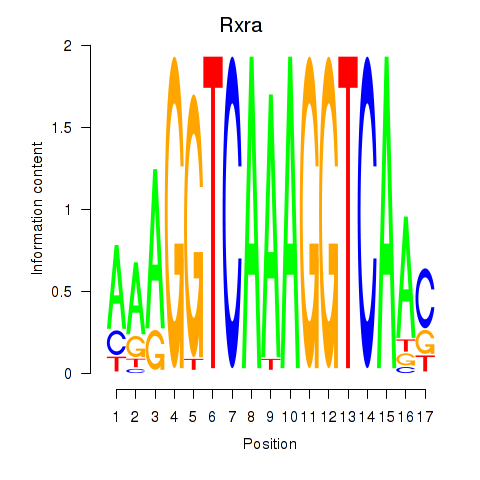
Transcription factors associated with Rxra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxra
|
ENSMUSG00000015846.16 | retinoid X receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxra | mm39_v1_chr2_+_27567213_27567241 | 0.98 | 2.3e-03 | Click! |
Activity profile of Rxra motif
Sorted Z-values of Rxra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_15135322 | 0.24 |
ENSMUST00000178312.2
|
Gm21897
|
predicted gene, 21897 |
| chr4_+_85123654 | 0.24 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr2_+_126057020 | 0.24 |
ENSMUST00000164042.3
|
Gm17555
|
predicted gene, 17555 |
| chr6_-_97408367 | 0.23 |
ENSMUST00000124050.3
|
Frmd4b
|
FERM domain containing 4B |
| chr9_-_110818679 | 0.22 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr11_-_11577824 | 0.21 |
ENSMUST00000081896.5
|
4930512M02Rik
|
RIKEN cDNA 4930512M02 gene |
| chr13_+_43519075 | 0.19 |
ENSMUST00000220458.2
ENSMUST00000220576.2 ENSMUST00000221515.2 |
Sirt5
|
sirtuin 5 |
| chr8_+_117822593 | 0.19 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr14_+_73898665 | 0.18 |
ENSMUST00000098874.4
|
Gm21750
|
predicted gene, 21750 |
| chr17_-_36220924 | 0.17 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr19_+_10502679 | 0.17 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr19_-_61215743 | 0.17 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr5_+_92719336 | 0.17 |
ENSMUST00000176621.8
ENSMUST00000175974.2 ENSMUST00000131166.9 ENSMUST00000176448.8 ENSMUST00000082382.8 |
Fam47e
|
family with sequence similarity 47, member E |
| chr12_-_113271532 | 0.16 |
ENSMUST00000192188.3
ENSMUST00000103418.3 |
Ighg2b
|
immunoglobulin heavy constant gamma 2B |
| chr13_+_43518927 | 0.16 |
ENSMUST00000221481.2
ENSMUST00000223194.2 |
Sirt5
|
sirtuin 5 |
| chr11_-_83483807 | 0.15 |
ENSMUST00000019071.4
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr7_+_110367375 | 0.15 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr16_+_11140740 | 0.15 |
ENSMUST00000180792.8
|
Snx29
|
sorting nexin 29 |
| chr7_+_140239843 | 0.14 |
ENSMUST00000218865.2
|
Olfr539
|
olfactory receptor 539 |
| chr19_-_45986919 | 0.14 |
ENSMUST00000045396.9
|
Armh3
|
armadillo-like helical domain containing 3 |
| chr2_-_58990967 | 0.14 |
ENSMUST00000226455.2
ENSMUST00000077687.6 |
Ccdc148
|
coiled-coil domain containing 148 |
| chrX_+_10351360 | 0.14 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr6_+_136509922 | 0.13 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_-_74544946 | 0.13 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr7_-_98305737 | 0.13 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr11_-_33463627 | 0.13 |
ENSMUST00000037522.14
|
Ranbp17
|
RAN binding protein 17 |
| chr16_+_11140779 | 0.12 |
ENSMUST00000115814.4
|
Snx29
|
sorting nexin 29 |
| chr19_-_12302465 | 0.12 |
ENSMUST00000207241.3
|
Olfr1437
|
olfactory receptor 1437 |
| chr11_+_75422516 | 0.12 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr4_-_103072343 | 0.12 |
ENSMUST00000150285.8
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr7_-_103535459 | 0.12 |
ENSMUST00000216303.2
|
Olfr66
|
olfactory receptor 66 |
| chr7_-_98305986 | 0.12 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr3_+_95041399 | 0.12 |
ENSMUST00000066386.6
|
Lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr3_-_95041246 | 0.11 |
ENSMUST00000172572.9
ENSMUST00000173462.3 |
Scnm1
|
sodium channel modifier 1 |
| chr7_-_30259025 | 0.11 |
ENSMUST00000043975.11
ENSMUST00000156241.2 |
Lin37
|
lin-37 homolog (C. elegans) |
| chr2_-_165210622 | 0.11 |
ENSMUST00000141140.2
ENSMUST00000103085.8 |
Zfp663
|
zinc finger protein 663 |
| chr14_-_52277310 | 0.11 |
ENSMUST00000216907.2
ENSMUST00000214071.2 ENSMUST00000216188.2 |
Olfr221
|
olfactory receptor 221 |
| chr11_+_97206542 | 0.10 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr2_+_70948267 | 0.10 |
ENSMUST00000028403.3
|
Cybrd1
|
cytochrome b reductase 1 |
| chr7_+_55417967 | 0.10 |
ENSMUST00000060416.15
ENSMUST00000094360.13 ENSMUST00000165045.9 ENSMUST00000173835.2 |
Siglech
|
sialic acid binding Ig-like lectin H |
| chr5_-_143831842 | 0.10 |
ENSMUST00000079624.12
ENSMUST00000110717.9 |
Ankrd61
|
ankyrin repeat domain 61 |
| chr7_-_18266150 | 0.10 |
ENSMUST00000094795.5
|
Psg25
|
pregnancy-specific glycoprotein 25 |
| chr2_+_172994841 | 0.10 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr3_-_102871440 | 0.09 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr4_+_118560720 | 0.09 |
ENSMUST00000214131.2
ENSMUST00000215117.3 |
Olfr1341
|
olfactory receptor 1341 |
| chr7_-_30259253 | 0.09 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr5_+_31107390 | 0.09 |
ENSMUST00000006814.9
|
Abhd1
|
abhydrolase domain containing 1 |
| chr13_+_13764982 | 0.09 |
ENSMUST00000110559.3
|
Lyst
|
lysosomal trafficking regulator |
| chr17_-_36220518 | 0.09 |
ENSMUST00000141132.2
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr9_+_44684248 | 0.08 |
ENSMUST00000128150.8
|
Ift46
|
intraflagellar transport 46 |
| chr1_-_170133901 | 0.08 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694 |
| chr8_-_46452896 | 0.08 |
ENSMUST00000053558.10
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr15_-_77331660 | 0.07 |
ENSMUST00000089469.7
|
Apol7b
|
apolipoprotein L 7b |
| chr2_+_157870399 | 0.07 |
ENSMUST00000103123.10
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr7_-_19411866 | 0.07 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr7_-_28913382 | 0.07 |
ENSMUST00000169143.8
ENSMUST00000047846.13 |
Catsperg1
|
cation channel sperm associated auxiliary subunit gamma 1 |
| chr10_+_62897353 | 0.07 |
ENSMUST00000178684.3
ENSMUST00000020266.15 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr6_+_21985902 | 0.07 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr9_+_108337726 | 0.07 |
ENSMUST00000061209.7
ENSMUST00000193269.2 |
Ccdc71
|
coiled-coil domain containing 71 |
| chr11_-_99134885 | 0.07 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr15_+_77583104 | 0.07 |
ENSMUST00000096358.6
|
Apol7e
|
apolipoprotein L 7e |
| chr2_+_88644840 | 0.06 |
ENSMUST00000214703.2
|
Olfr1202
|
olfactory receptor 1202 |
| chr7_-_19684654 | 0.06 |
ENSMUST00000043440.8
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chr2_+_172235702 | 0.06 |
ENSMUST00000099061.9
ENSMUST00000103073.9 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr5_-_109704339 | 0.06 |
ENSMUST00000198960.2
|
Crlf2
|
cytokine receptor-like factor 2 |
| chrX_-_41000746 | 0.06 |
ENSMUST00000047037.15
|
Thoc2
|
THO complex 2 |
| chr4_+_126450762 | 0.06 |
ENSMUST00000147675.2
|
Clspn
|
claspin |
| chr9_-_44632680 | 0.05 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr9_-_78285942 | 0.05 |
ENSMUST00000034900.8
|
Ooep
|
oocyte expressed protein |
| chr6_+_68279392 | 0.05 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr5_+_136023649 | 0.05 |
ENSMUST00000111142.9
ENSMUST00000111145.10 ENSMUST00000111144.8 ENSMUST00000199239.5 ENSMUST00000005072.10 ENSMUST00000130345.2 |
Dtx2
|
deltex 2, E3 ubiquitin ligase |
| chr4_+_135622694 | 0.05 |
ENSMUST00000097843.9
|
Cnr2
|
cannabinoid receptor 2 (macrophage) |
| chr4_-_118266416 | 0.05 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr9_+_44684285 | 0.05 |
ENSMUST00000239014.2
|
Ift46
|
intraflagellar transport 46 |
| chrX_+_158197568 | 0.05 |
ENSMUST00000112471.9
|
Map7d2
|
MAP7 domain containing 2 |
| chr9_-_44876817 | 0.04 |
ENSMUST00000214761.2
ENSMUST00000213666.2 ENSMUST00000213890.2 ENSMUST00000125642.8 ENSMUST00000213193.2 ENSMUST00000117506.9 ENSMUST00000138559.9 ENSMUST00000117549.8 |
Ube4a
|
ubiquitination factor E4A |
| chr7_+_55417921 | 0.04 |
ENSMUST00000121492.8
ENSMUST00000171077.8 |
Siglech
|
sialic acid binding Ig-like lectin H |
| chrX_+_99537897 | 0.04 |
ENSMUST00000033570.6
|
Igbp1
|
immunoglobulin (CD79A) binding protein 1 |
| chr17_+_29579882 | 0.04 |
ENSMUST00000024810.8
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr11_-_116089866 | 0.04 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr7_+_132460954 | 0.04 |
ENSMUST00000084497.12
ENSMUST00000106161.8 |
Abraxas2
|
BRISC complex subunit |
| chr4_+_97665992 | 0.04 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr9_+_44684206 | 0.04 |
ENSMUST00000002099.11
|
Ift46
|
intraflagellar transport 46 |
| chr16_+_33338648 | 0.04 |
ENSMUST00000122427.8
ENSMUST00000059056.15 |
Slc12a8
|
solute carrier family 12 (potassium/chloride transporters), member 8 |
| chr2_+_37029334 | 0.04 |
ENSMUST00000214905.2
ENSMUST00000217298.2 ENSMUST00000104995.3 |
Olfr364-ps1
|
olfactory receptor 364, pseudogene 1 |
| chr9_-_109168710 | 0.03 |
ENSMUST00000196351.2
ENSMUST00000200156.5 ENSMUST00000112040.9 ENSMUST00000112039.7 |
Fbxw28
|
F-box and WD-40 domain protein 28 |
| chr2_-_157870341 | 0.03 |
ENSMUST00000029179.11
|
Tti1
|
TELO2 interacting protein 1 |
| chr8_+_88978600 | 0.03 |
ENSMUST00000154115.2
|
Tent4b
|
terminal nucleotidyltransferase 4B |
| chr9_-_108338111 | 0.03 |
ENSMUST00000193895.6
|
Klhdc8b
|
kelch domain containing 8B |
| chr6_-_116561156 | 0.03 |
ENSMUST00000061723.6
|
Olfr215
|
olfactory receptor 215 |
| chr2_+_157870606 | 0.03 |
ENSMUST00000109518.8
ENSMUST00000029180.14 |
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr9_-_48876290 | 0.03 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr16_+_4825170 | 0.03 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chr13_-_112717196 | 0.03 |
ENSMUST00000051756.8
|
Il31ra
|
interleukin 31 receptor A |
| chr1_+_63215976 | 0.03 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr7_+_18946448 | 0.03 |
ENSMUST00000060225.6
|
Gpr4
|
G protein-coupled receptor 4 |
| chr9_+_118335294 | 0.03 |
ENSMUST00000084820.6
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr14_+_12016304 | 0.03 |
ENSMUST00000161302.8
|
Fhit
|
fragile histidine triad gene |
| chr5_-_73132045 | 0.03 |
ENSMUST00000043711.9
|
Gm10135
|
predicted gene 10135 |
| chr9_+_20914012 | 0.03 |
ENSMUST00000003386.7
|
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr8_+_71921824 | 0.03 |
ENSMUST00000124745.8
ENSMUST00000138892.2 ENSMUST00000147642.2 |
Dda1
|
DET1 and DDB1 associated 1 |
| chrX_+_100492684 | 0.03 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr5_+_76988444 | 0.03 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr5_+_31855304 | 0.03 |
ENSMUST00000114515.9
|
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr2_+_157870653 | 0.03 |
ENSMUST00000152452.8
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr12_-_115876396 | 0.03 |
ENSMUST00000103547.2
|
Ighv1-80
|
immunoglobulin heavy variable 1-80 |
| chr8_-_112660363 | 0.02 |
ENSMUST00000034429.9
|
Tmem231
|
transmembrane protein 231 |
| chr15_+_79230777 | 0.02 |
ENSMUST00000229130.2
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr11_-_116089595 | 0.02 |
ENSMUST00000072948.11
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr2_+_121188195 | 0.02 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr10_+_74896383 | 0.02 |
ENSMUST00000164107.3
|
Bcr
|
BCR activator of RhoGEF and GTPase |
| chr14_+_73380163 | 0.02 |
ENSMUST00000170368.8
ENSMUST00000171767.8 ENSMUST00000163533.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr17_-_35351026 | 0.02 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr7_+_98143429 | 0.02 |
ENSMUST00000165205.2
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr11_-_53313950 | 0.02 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr5_+_31855394 | 0.02 |
ENSMUST00000063813.11
ENSMUST00000071531.12 ENSMUST00000131995.7 ENSMUST00000114507.10 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr16_+_4825146 | 0.02 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr11_-_118233326 | 0.02 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr4_-_99717359 | 0.02 |
ENSMUST00000146258.2
|
Itgb3bp
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr15_+_86098660 | 0.02 |
ENSMUST00000063414.9
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr16_+_4825216 | 0.02 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chrX_+_100576340 | 0.02 |
ENSMUST00000118878.8
ENSMUST00000101341.9 ENSMUST00000149274.9 |
Taf1
|
TATA-box binding protein associated factor 1 |
| chr6_-_85797917 | 0.02 |
ENSMUST00000174143.2
|
Nat8f6
|
N-acetyltransferase 8 (GCN5-related) family member 6 |
| chr10_-_76459152 | 0.02 |
ENSMUST00000105413.8
|
Col6a2
|
collagen, type VI, alpha 2 |
| chr13_-_52685305 | 0.02 |
ENSMUST00000057442.8
|
Diras2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr14_+_67982630 | 0.02 |
ENSMUST00000223929.2
ENSMUST00000111095.4 |
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr19_+_9979033 | 0.02 |
ENSMUST00000121418.8
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr2_+_155453103 | 0.01 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chrX_-_147398149 | 0.01 |
ENSMUST00000112755.3
|
Gm8334
|
predicted gene 8334 |
| chr3_+_107784543 | 0.01 |
ENSMUST00000037375.10
ENSMUST00000199990.2 |
Eps8l3
|
EPS8-like 3 |
| chr14_+_54032814 | 0.01 |
ENSMUST00000103671.4
|
Trav13-5
|
T cell receptor alpha variable 13-5 |
| chr18_+_63841756 | 0.01 |
ENSMUST00000072726.7
ENSMUST00000235648.2 ENSMUST00000236879.2 |
Wdr7
|
WD repeat domain 7 |
| chr9_+_108269992 | 0.01 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr11_-_100650768 | 0.01 |
ENSMUST00000107363.3
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr7_-_43906629 | 0.01 |
ENSMUST00000012921.9
|
Acp4
|
acid phosphatase 4 |
| chr9_+_45014092 | 0.01 |
ENSMUST00000217074.2
|
Jaml
|
junction adhesion molecule like |
| chr11_+_85202058 | 0.01 |
ENSMUST00000020835.16
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr1_-_132635042 | 0.01 |
ENSMUST00000043189.14
|
Nfasc
|
neurofascin |
| chr11_-_86999481 | 0.01 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr2_-_32665596 | 0.01 |
ENSMUST00000161430.8
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr14_+_122116002 | 0.01 |
ENSMUST00000039803.7
|
Ubac2
|
ubiquitin associated domain containing 2 |
| chr9_-_108526548 | 0.01 |
ENSMUST00000013338.14
ENSMUST00000193197.6 |
Arih2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
| chr2_-_19558719 | 0.01 |
ENSMUST00000062060.5
|
4921504E06Rik
|
RIKEN cDNA 4921504E06 gene |
| chr4_+_126450728 | 0.01 |
ENSMUST00000048391.15
|
Clspn
|
claspin |
| chr12_+_112455882 | 0.01 |
ENSMUST00000057465.7
ENSMUST00000223266.2 |
A530016L24Rik
|
RIKEN cDNA A530016L24 gene |
| chr9_+_108270020 | 0.01 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr15_-_98155173 | 0.01 |
ENSMUST00000060855.7
|
H1f7
|
H1.7 linker histone |
| chr10_+_14581325 | 0.01 |
ENSMUST00000191238.7
ENSMUST00000190114.2 |
Nmbr
|
neuromedin B receptor |
| chr11_+_121150798 | 0.00 |
ENSMUST00000106113.2
|
Foxk2
|
forkhead box K2 |
| chr10_-_82459024 | 0.00 |
ENSMUST00000183363.2
ENSMUST00000079648.12 ENSMUST00000185168.8 |
1190007I07Rik
|
RIKEN cDNA 1190007I07 gene |
| chr1_-_84912810 | 0.00 |
ENSMUST00000027422.7
|
Slc16a14
|
solute carrier family 16 (monocarboxylic acid transporters), member 14 |
| chr17_+_49922129 | 0.00 |
ENSMUST00000162854.2
|
Kif6
|
kinesin family member 6 |
| chr1_+_190769010 | 0.00 |
ENSMUST00000077889.8
|
Spata45
|
spermatogenesis associated 45 |
| chr19_-_3962733 | 0.00 |
ENSMUST00000075092.8
ENSMUST00000235847.2 ENSMUST00000235301.2 ENSMUST00000237341.2 |
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr6_+_8259328 | 0.00 |
ENSMUST00000159378.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr17_-_15784582 | 0.00 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
| chr10_+_119655294 | 0.00 |
ENSMUST00000105262.9
ENSMUST00000147454.8 ENSMUST00000138410.8 ENSMUST00000144825.8 ENSMUST00000148954.8 ENSMUST00000144959.8 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr13_+_22574543 | 0.00 |
ENSMUST00000226157.2
ENSMUST00000227326.2 ENSMUST00000228726.2 |
Vmn1r200
|
vomeronasal 1 receptor 200 |
| chr5_+_145282064 | 0.00 |
ENSMUST00000079268.9
|
Cyp3a57
|
cytochrome P450, family 3, subfamily a, polypeptide 57 |
| chr14_+_69409251 | 0.00 |
ENSMUST00000062437.10
|
Nkx2-6
|
NK2 homeobox 6 |
| chr14_+_106343614 | 0.00 |
ENSMUST00000022708.7
|
Trim52
|
tripartite motif-containing 52 |
| chr6_+_42716305 | 0.00 |
ENSMUST00000213997.2
|
Olfr453
|
olfactory receptor 453 |
| chr1_+_74448535 | 0.00 |
ENSMUST00000027366.13
|
Vil1
|
villin 1 |
| chr15_+_89417017 | 0.00 |
ENSMUST00000167173.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr2_-_119060330 | 0.00 |
ENSMUST00000110820.3
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr11_+_50938936 | 0.00 |
ENSMUST00000073543.2
|
Olfr1375
|
olfactory receptor 1375 |
| chr9_+_40059408 | 0.00 |
ENSMUST00000046333.9
ENSMUST00000238613.2 |
Tmem225
|
transmembrane protein 225 |
| chr3_+_36530081 | 0.00 |
ENSMUST00000029268.7
|
1810062G17Rik
|
RIKEN cDNA 1810062G17 gene |
| chr8_-_86107593 | 0.00 |
ENSMUST00000122452.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr12_+_83810402 | 0.00 |
ENSMUST00000121733.8
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr19_-_11848669 | 0.00 |
ENSMUST00000087857.3
|
Olfr1419
|
olfactory receptor 1419 |
| chr7_+_26821266 | 0.00 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr8_-_100143029 | 0.00 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr2_-_3420056 | 0.00 |
ENSMUST00000115084.8
ENSMUST00000115083.8 |
Meig1
|
meiosis expressed gene 1 |
| chr9_-_103165489 | 0.00 |
ENSMUST00000035163.10
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr4_+_117581609 | 0.00 |
ENSMUST00000203629.2
ENSMUST00000204276.2 |
Gm12845
|
predicted gene 12845 |
| chr4_+_41569775 | 0.00 |
ENSMUST00000102963.10
|
Dnai1
|
dynein axonemal intermediate chain 1 |
| chr12_-_103706774 | 0.00 |
ENSMUST00000186166.7
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr4_-_137301880 | 0.00 |
ENSMUST00000178923.2
|
Ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr2_-_3420102 | 0.00 |
ENSMUST00000115082.10
|
Meig1
|
meiosis expressed gene 1 |
| chr14_+_53903370 | 0.00 |
ENSMUST00000181768.3
|
Trav3-3
|
T cell receptor alpha variable 3-3 |
| chr9_-_103165423 | 0.00 |
ENSMUST00000123530.8
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr14_+_53210216 | 0.00 |
ENSMUST00000103599.3
|
Trav3d-3
|
T cell receptor alpha variable 3D-3 |
| chr2_-_32665637 | 0.00 |
ENSMUST00000161958.2
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr2_-_121786573 | 0.00 |
ENSMUST00000104936.4
|
Mageb3
|
MAGE family member B3 |
| chr6_+_68495964 | 0.00 |
ENSMUST00000199510.5
ENSMUST00000103325.3 |
Igkv14-100
|
immunoglobulin kappa chain variable 14-100 |
| chr14_-_14512458 | 0.00 |
ENSMUST00000211339.2
|
Gm45521
|
predicted gene 45521 |
| chrX_+_36518660 | 0.00 |
ENSMUST00000185028.8
ENSMUST00000185050.3 |
Rhox3a
|
reproductive homeobox 3A |
| chrX_-_36485911 | 0.00 |
ENSMUST00000115210.2
|
Rhox1
|
reproductive homeobox 1 |
| chr6_-_85797946 | 0.00 |
ENSMUST00000032074.5
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr7_+_103209686 | 0.00 |
ENSMUST00000098198.3
|
Olfr615
|
olfactory receptor 615 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.1 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.0 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |