Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Rxrb
Z-value: 0.61
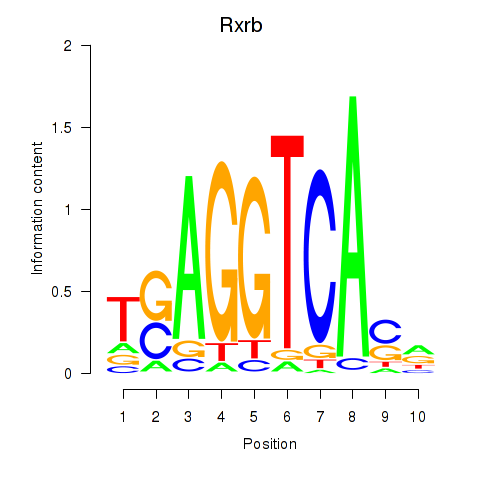
Transcription factors associated with Rxrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxrb
|
ENSMUSG00000039656.18 | retinoid X receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrb | mm39_v1_chr17_+_34251322_34251381 | -0.94 | 1.6e-02 | Click! |
Activity profile of Rxrb motif
Sorted Z-values of Rxrb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_140111138 | 0.83 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 0.74 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr17_+_21503204 | 0.56 |
ENSMUST00000236545.2
ENSMUST00000235527.2 |
Vmn1r236
|
vomeronasal 1 receptor 236 |
| chr12_+_33364288 | 0.39 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr15_-_5273659 | 0.34 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr15_-_5273645 | 0.33 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr7_+_3339059 | 0.31 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_+_3339077 | 0.30 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr16_+_24540599 | 0.29 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_+_71884943 | 0.29 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr8_+_72860884 | 0.26 |
ENSMUST00000210435.4
|
Olfr374
|
olfactory receptor 374 |
| chr10_-_67972401 | 0.24 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chrX_+_72760183 | 0.23 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr7_-_10292412 | 0.22 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr11_-_97591150 | 0.19 |
ENSMUST00000018681.14
|
Pcgf2
|
polycomb group ring finger 2 |
| chr8_-_71245036 | 0.18 |
ENSMUST00000212038.2
ENSMUST00000212551.2 ENSMUST00000211948.2 |
Mast3
|
microtubule associated serine/threonine kinase 3 |
| chr1_+_172327569 | 0.18 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr6_-_124942457 | 0.17 |
ENSMUST00000112439.9
|
Cops7a
|
COP9 signalosome subunit 7A |
| chr11_-_104332528 | 0.17 |
ENSMUST00000106971.2
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr13_+_44883270 | 0.17 |
ENSMUST00000172830.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr11_-_97590460 | 0.17 |
ENSMUST00000103148.8
ENSMUST00000169807.8 |
Pcgf2
|
polycomb group ring finger 2 |
| chr6_+_136509922 | 0.17 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr4_-_62278761 | 0.16 |
ENSMUST00000107461.2
ENSMUST00000084528.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr12_+_111679689 | 0.15 |
ENSMUST00000040519.12
ENSMUST00000163220.10 ENSMUST00000162316.2 |
Coa8
|
cytochrome c oxidase assembly factor 8 |
| chr7_-_26317889 | 0.15 |
ENSMUST00000228467.2
ENSMUST00000227479.2 ENSMUST00000228367.2 ENSMUST00000228004.2 ENSMUST00000227695.2 ENSMUST00000228133.2 ENSMUST00000227264.2 ENSMUST00000228676.2 ENSMUST00000226694.2 ENSMUST00000227461.2 ENSMUST00000227411.2 ENSMUST00000228633.2 |
Vmn1r185
|
vomeronasal 1 receptor 185 |
| chr14_+_58308004 | 0.15 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr5_-_137623331 | 0.14 |
ENSMUST00000031732.14
|
Fbxo24
|
F-box protein 24 |
| chr6_-_124942366 | 0.14 |
ENSMUST00000129446.8
ENSMUST00000032220.15 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr14_+_47710574 | 0.14 |
ENSMUST00000228740.2
|
Fbxo34
|
F-box protein 34 |
| chr4_-_139695337 | 0.13 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr17_+_48571298 | 0.13 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr4_-_41048124 | 0.13 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr6_+_22288220 | 0.13 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr2_+_112096154 | 0.12 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr3_-_94693740 | 0.12 |
ENSMUST00000153263.9
ENSMUST00000107272.7 ENSMUST00000155485.4 |
Cgn
|
cingulin |
| chr5_+_73071897 | 0.12 |
ENSMUST00000200785.2
|
Slain2
|
SLAIN motif family, member 2 |
| chr11_-_70146156 | 0.12 |
ENSMUST00000108574.3
ENSMUST00000000329.9 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr11_-_74480870 | 0.12 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chrX_+_10351360 | 0.11 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr10_+_79984097 | 0.11 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chr3_+_96079642 | 0.11 |
ENSMUST00000076372.5
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr16_-_23339548 | 0.11 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr2_+_48839505 | 0.11 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr4_-_41275091 | 0.10 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr13_+_43276323 | 0.10 |
ENSMUST00000136576.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr19_-_6957789 | 0.10 |
ENSMUST00000070878.9
ENSMUST00000177752.9 |
Fkbp2
|
FK506 binding protein 2 |
| chr1_-_38168801 | 0.10 |
ENSMUST00000195383.2
|
Rev1
|
REV1, DNA directed polymerase |
| chr11_+_100960838 | 0.09 |
ENSMUST00000001802.10
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| chrX_-_48823936 | 0.09 |
ENSMUST00000215373.3
|
Olfr1321
|
olfactory receptor 1321 |
| chr15_-_82238432 | 0.09 |
ENSMUST00000230494.2
ENSMUST00000023085.7 |
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr7_+_28140352 | 0.09 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr3_-_94693780 | 0.08 |
ENSMUST00000107273.9
ENSMUST00000238849.2 |
Cgn
|
cingulin |
| chr4_-_20778847 | 0.08 |
ENSMUST00000102998.4
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr11_+_101875095 | 0.08 |
ENSMUST00000176722.8
ENSMUST00000175972.2 |
Cfap97d1
|
CFAP97 domain containing 1 |
| chr14_-_52277310 | 0.08 |
ENSMUST00000216907.2
ENSMUST00000214071.2 ENSMUST00000216188.2 |
Olfr221
|
olfactory receptor 221 |
| chr4_+_152262583 | 0.08 |
ENSMUST00000075363.10
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr19_+_11863929 | 0.08 |
ENSMUST00000217281.2
|
Olfr1420
|
olfactory receptor 1420 |
| chr19_+_42040681 | 0.08 |
ENSMUST00000164518.4
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
| chr11_-_101875575 | 0.08 |
ENSMUST00000176261.2
ENSMUST00000143177.2 ENSMUST00000003612.13 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr3_+_122522592 | 0.08 |
ENSMUST00000066728.10
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr11_+_97206542 | 0.07 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr3_-_89300936 | 0.07 |
ENSMUST00000124783.8
ENSMUST00000126027.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr10_-_8761777 | 0.07 |
ENSMUST00000015449.6
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr2_-_79959178 | 0.06 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_+_121279842 | 0.06 |
ENSMUST00000110615.8
ENSMUST00000099475.12 |
Serf2
|
small EDRK-rich factor 2 |
| chr6_+_71520855 | 0.06 |
ENSMUST00000204535.2
ENSMUST00000065364.5 ENSMUST00000204199.2 |
Chmp3
|
charged multivesicular body protein 3 |
| chr2_-_6726417 | 0.06 |
ENSMUST00000142941.8
ENSMUST00000150624.9 ENSMUST00000100429.11 ENSMUST00000182879.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr11_-_118233326 | 0.06 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr1_+_159871943 | 0.06 |
ENSMUST00000163892.8
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene |
| chr5_+_73071695 | 0.05 |
ENSMUST00000143829.5
|
Slain2
|
SLAIN motif family, member 2 |
| chr2_+_25132941 | 0.05 |
ENSMUST00000114355.2
ENSMUST00000060818.2 |
Rnf208
|
ring finger protein 208 |
| chr11_-_95037089 | 0.05 |
ENSMUST00000021241.8
|
Dlx4
|
distal-less homeobox 4 |
| chr1_-_162305573 | 0.05 |
ENSMUST00000086074.12
ENSMUST00000070330.14 |
Dnm3
|
dynamin 3 |
| chr11_-_53321606 | 0.05 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr5_-_123804745 | 0.04 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr17_+_25097199 | 0.04 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr14_-_55344004 | 0.04 |
ENSMUST00000036041.15
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chrX_+_98086187 | 0.04 |
ENSMUST00000036606.14
|
Stard8
|
START domain containing 8 |
| chr15_+_76152276 | 0.04 |
ENSMUST00000074173.4
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr6_+_129510145 | 0.04 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr2_-_25511936 | 0.04 |
ENSMUST00000028308.11
ENSMUST00000142087.2 |
Tmem141
|
transmembrane protein 141 |
| chr4_+_57845240 | 0.04 |
ENSMUST00000102903.8
ENSMUST00000107598.9 |
Pakap
|
paralemmin A kinase anchor protein |
| chr17_-_24863956 | 0.04 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr9_-_106769069 | 0.04 |
ENSMUST00000160503.4
ENSMUST00000159620.9 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr11_-_99134885 | 0.04 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr17_-_24863907 | 0.04 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr17_-_37178079 | 0.03 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr7_+_15909283 | 0.03 |
ENSMUST00000002495.18
|
Meis3
|
Meis homeobox 3 |
| chr17_-_71575584 | 0.03 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr14_+_6226418 | 0.03 |
ENSMUST00000112625.9
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr8_-_106732607 | 0.03 |
ENSMUST00000227778.2
|
Dpep2nb
|
Dpep2 neighbor |
| chr6_-_124942170 | 0.03 |
ENSMUST00000148485.2
ENSMUST00000129976.8 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr7_+_16693604 | 0.03 |
ENSMUST00000038163.8
|
Pnmal1
|
PNMA-like 1 |
| chr5_+_110987839 | 0.03 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr7_+_15909010 | 0.03 |
ENSMUST00000176342.8
ENSMUST00000177540.8 |
Meis3
|
Meis homeobox 3 |
| chr5_-_25200745 | 0.02 |
ENSMUST00000076306.12
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr19_-_6036171 | 0.02 |
ENSMUST00000041827.8
|
Slc22a20
|
solute carrier family 22 (organic anion transporter), member 20 |
| chr8_-_13612397 | 0.02 |
ENSMUST00000187391.7
ENSMUST00000134023.9 ENSMUST00000151400.10 |
1700029H14Rik
|
RIKEN cDNA 1700029H14 gene |
| chr9_-_106769131 | 0.02 |
ENSMUST00000159283.8
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr2_-_150934270 | 0.02 |
ENSMUST00000109890.2
|
Gm14151
|
predicted gene 14151 |
| chr7_+_40636967 | 0.02 |
ENSMUST00000206529.2
ENSMUST00000171664.2 |
4930433I11Rik
|
RIKEN cDNA 4930433I11 gene |
| chr7_+_105289655 | 0.02 |
ENSMUST00000058333.10
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr15_-_73702827 | 0.02 |
ENSMUST00000238402.2
|
Mroh5
|
maestro heat-like repeat family member 5 |
| chr6_+_119213468 | 0.02 |
ENSMUST00000238905.2
ENSMUST00000037434.13 |
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr2_-_151318073 | 0.02 |
ENSMUST00000080132.3
|
4921509C19Rik
|
RIKEN cDNA 4921509C19 gene |
| chr1_-_170133901 | 0.02 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694 |
| chr6_-_68812291 | 0.02 |
ENSMUST00000199143.5
ENSMUST00000103335.3 |
Igkv12-89
|
immunoglobulin kappa chain variable 12-89 |
| chr5_+_124065481 | 0.02 |
ENSMUST00000166129.5
|
Gm43518
|
predicted gene 43518 |
| chr5_-_110987604 | 0.02 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr11_-_40624200 | 0.02 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr17_-_18075450 | 0.02 |
ENSMUST00000003762.8
|
Has1
|
hyaluronan synthase 1 |
| chr16_+_20513658 | 0.02 |
ENSMUST00000056518.13
|
Fam131a
|
family with sequence similarity 131, member A |
| chr1_+_133109059 | 0.01 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr4_-_106585127 | 0.01 |
ENSMUST00000106770.8
ENSMUST00000145044.2 |
Mroh7
|
maestro heat-like repeat family member 7 |
| chr3_+_38941089 | 0.01 |
ENSMUST00000061260.8
|
Fat4
|
FAT atypical cadherin 4 |
| chr12_-_113589576 | 0.01 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chrX_+_85235370 | 0.01 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr1_-_38168697 | 0.01 |
ENSMUST00000027251.12
|
Rev1
|
REV1, DNA directed polymerase |
| chr17_+_28794615 | 0.01 |
ENSMUST00000232862.2
ENSMUST00000080780.8 |
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr1_+_54369151 | 0.01 |
ENSMUST00000163072.8
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr7_-_10011933 | 0.01 |
ENSMUST00000227719.2
ENSMUST00000228622.2 ENSMUST00000228086.2 |
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr4_+_110254858 | 0.01 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr6_+_96092230 | 0.01 |
ENSMUST00000075080.6
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr4_+_110254907 | 0.01 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr12_+_91367764 | 0.01 |
ENSMUST00000021346.14
ENSMUST00000021343.8 |
Tshr
|
thyroid stimulating hormone receptor |
| chr19_+_8861274 | 0.01 |
ENSMUST00000177826.2
|
Gm49403
|
predicted gene, 49403 |
| chr16_-_44153498 | 0.01 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr9_+_55116209 | 0.01 |
ENSMUST00000034859.15
|
Fbxo22
|
F-box protein 22 |
| chr2_-_122144125 | 0.01 |
ENSMUST00000147788.8
ENSMUST00000154412.2 ENSMUST00000110537.8 ENSMUST00000148417.8 |
Duoxa1
|
dual oxidase maturation factor 1 |
| chr12_+_86288626 | 0.01 |
ENSMUST00000221368.2
ENSMUST00000071106.6 |
Gpatch2l
|
G patch domain containing 2 like |
| chr5_-_74863514 | 0.01 |
ENSMUST00000117388.8
|
Lnx1
|
ligand of numb-protein X 1 |
| chr11_-_94376158 | 0.01 |
ENSMUST00000041705.8
|
Spata20
|
spermatogenesis associated 20 |
| chr11_-_3672188 | 0.01 |
ENSMUST00000102950.10
ENSMUST00000101632.4 |
Osbp2
|
oxysterol binding protein 2 |
| chr1_+_133291302 | 0.01 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr14_+_54713703 | 0.01 |
ENSMUST00000164697.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr2_-_6217844 | 0.01 |
ENSMUST00000042658.5
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr7_-_104835219 | 0.01 |
ENSMUST00000214712.2
ENSMUST00000217432.2 ENSMUST00000209409.3 |
Olfr685
|
olfactory receptor 685 |
| chr2_+_150940165 | 0.01 |
ENSMUST00000109888.2
|
Gm14147
|
predicted gene 14147 |
| chr16_-_44153288 | 0.01 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr7_-_4774277 | 0.01 |
ENSMUST00000174409.2
|
Fam71e2
|
family with sequence similarity 71, member E2 |
| chr9_+_92339422 | 0.01 |
ENSMUST00000034941.9
|
Plscr4
|
phospholipid scramblase 4 |
| chr18_+_35987791 | 0.01 |
ENSMUST00000235404.2
|
Cxxc5
|
CXXC finger 5 |
| chr3_+_107009896 | 0.01 |
ENSMUST00000196403.2
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr18_+_57275854 | 0.01 |
ENSMUST00000139892.2
|
Megf10
|
multiple EGF-like-domains 10 |
| chr15_+_92059224 | 0.01 |
ENSMUST00000068378.6
|
Cntn1
|
contactin 1 |
| chr7_+_45164005 | 0.01 |
ENSMUST00000210532.2
ENSMUST00000085331.14 ENSMUST00000210299.2 |
Tulp2
|
tubby-like protein 2 |
| chr8_+_77626400 | 0.01 |
ENSMUST00000109913.9
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr2_-_25104628 | 0.01 |
ENSMUST00000043774.11
ENSMUST00000114363.2 |
Stpg3
|
sperm tail PG rich repeat containing 3 |
| chr1_-_156936197 | 0.01 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr2_-_136229849 | 0.01 |
ENSMUST00000035264.9
ENSMUST00000077200.4 |
Pak5
|
p21 (RAC1) activated kinase 5 |
| chr14_-_21791544 | 0.01 |
ENSMUST00000184703.8
ENSMUST00000183698.8 ENSMUST00000119866.9 |
Dusp13
|
dual specificity phosphatase 13 |
| chr17_+_28794727 | 0.01 |
ENSMUST00000232974.2
|
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr6_+_125122172 | 0.01 |
ENSMUST00000119527.8
ENSMUST00000117675.8 |
Iffo1
|
intermediate filament family orphan 1 |
| chr1_+_75456173 | 0.01 |
ENSMUST00000113575.9
ENSMUST00000148980.2 ENSMUST00000050899.7 ENSMUST00000187411.2 |
Tmem198
|
transmembrane protein 198 |
| chr1_+_175526152 | 0.01 |
ENSMUST00000094288.10
ENSMUST00000171939.8 |
Wdr64
|
WD repeat domain 64 |
| chr2_-_153712996 | 0.01 |
ENSMUST00000028982.5
|
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr12_-_98703664 | 0.01 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr15_+_101345625 | 0.01 |
ENSMUST00000023781.9
|
Krt88
|
keratin 88 |
| chr7_-_30755007 | 0.01 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr16_-_10608766 | 0.01 |
ENSMUST00000050864.7
|
Prm3
|
protamine 3 |
| chr11_+_104077153 | 0.01 |
ENSMUST00000107000.2
ENSMUST00000059448.8 |
Sppl2c
|
signal peptide peptidase 2C |
| chr1_+_167445815 | 0.01 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr9_+_110595224 | 0.01 |
ENSMUST00000136695.3
|
Myl3
|
myosin, light polypeptide 3 |
| chr1_-_84817022 | 0.01 |
ENSMUST00000189496.7
ENSMUST00000027421.13 ENSMUST00000186894.7 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr7_-_119694400 | 0.01 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr5_-_116560916 | 0.01 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr5_+_73071607 | 0.00 |
ENSMUST00000144843.8
|
Slain2
|
SLAIN motif family, member 2 |
| chr11_+_117375194 | 0.00 |
ENSMUST00000092394.4
|
Gm11733
|
predicted gene 11733 |
| chr2_-_86470069 | 0.00 |
ENSMUST00000216851.2
|
Olfr1084
|
olfactory receptor 1084 |
| chr15_+_38219447 | 0.00 |
ENSMUST00000081966.5
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr4_-_20778530 | 0.00 |
ENSMUST00000119374.8
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr7_+_45164042 | 0.00 |
ENSMUST00000210868.2
|
Tulp2
|
tubby-like protein 2 |
| chr1_+_131678223 | 0.00 |
ENSMUST00000147800.2
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr1_-_13197387 | 0.00 |
ENSMUST00000047577.7
|
Prdm14
|
PR domain containing 14 |
| chr1_+_85949657 | 0.00 |
ENSMUST00000052854.13
ENSMUST00000125083.2 ENSMUST00000152501.8 ENSMUST00000113344.8 ENSMUST00000130504.8 ENSMUST00000153247.3 |
Spata3
|
spermatogenesis associated 3 |
| chr15_+_98953776 | 0.00 |
ENSMUST00000229268.2
|
Prph
|
peripherin |
| chr4_-_49597425 | 0.00 |
ENSMUST00000150664.2
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr3_+_122298308 | 0.00 |
ENSMUST00000199358.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr4_+_57821050 | 0.00 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr12_+_86781154 | 0.00 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr2_+_174602412 | 0.00 |
ENSMUST00000029030.9
|
Edn3
|
endothelin 3 |
| chr4_+_151081538 | 0.00 |
ENSMUST00000030803.2
|
Uts2
|
urotensin 2 |
| chr7_-_133966588 | 0.00 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr1_+_85949681 | 0.00 |
ENSMUST00000159876.8
ENSMUST00000135440.2 |
Spata3
|
spermatogenesis associated 3 |
| chr9_+_45313913 | 0.00 |
ENSMUST00000214257.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr13_-_59892731 | 0.00 |
ENSMUST00000180139.3
|
1700014D04Rik
|
RIKEN cDNA 1700014D04 gene |
| chr11_+_104983022 | 0.00 |
ENSMUST00000021029.6
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr3_-_59038539 | 0.00 |
ENSMUST00000198838.2
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr12_+_86781141 | 0.00 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr9_-_40915895 | 0.00 |
ENSMUST00000180384.3
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr9_-_40915858 | 0.00 |
ENSMUST00000188848.8
ENSMUST00000034519.13 |
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr9_+_38019984 | 0.00 |
ENSMUST00000075228.2
|
Olfr888
|
olfactory receptor 888 |
| chr14_-_118289557 | 0.00 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr7_-_105249308 | 0.00 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr19_-_7688628 | 0.00 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr2_-_140231618 | 0.00 |
ENSMUST00000122367.8
ENSMUST00000120133.2 |
Sel1l2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr2_+_174602574 | 0.00 |
ENSMUST00000140908.2
|
Edn3
|
endothelin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxrb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033624 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.2 | 1.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 | 0.1 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |