Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Scrt2
Z-value: 0.86
Transcription factors associated with Scrt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt2
|
ENSMUSG00000060257.3 | scratch family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt2 | mm39_v1_chr2_+_151923449_151923544 | 0.84 | 7.3e-02 | Click! |
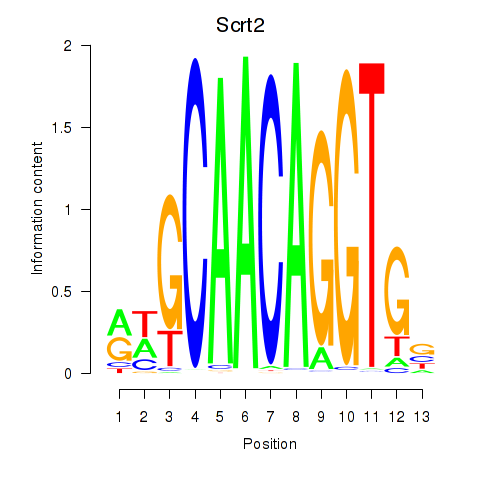
Activity profile of Scrt2 motif
Sorted Z-values of Scrt2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_71318353 | 1.47 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr13_+_23940964 | 1.14 |
ENSMUST00000102965.4
|
H4c2
|
H4 clustered histone 2 |
| chr3_-_122778052 | 0.64 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr1_-_156767123 | 0.63 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr8_+_91635192 | 0.58 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr13_+_109397184 | 0.50 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr1_+_182236728 | 0.47 |
ENSMUST00000117245.2
|
Trp53bp2
|
transformation related protein 53 binding protein 2 |
| chrX_-_132882514 | 0.47 |
ENSMUST00000113297.9
ENSMUST00000174542.2 ENSMUST00000033608.15 ENSMUST00000113294.8 |
Sytl4
|
synaptotagmin-like 4 |
| chr4_-_155947819 | 0.45 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr2_-_60711706 | 0.45 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr16_+_16888145 | 0.43 |
ENSMUST00000232574.2
|
Ypel1
|
yippee like 1 |
| chr2_-_168432111 | 0.42 |
ENSMUST00000029057.13
ENSMUST00000074618.10 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr4_-_135221810 | 0.35 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr16_+_3702493 | 0.34 |
ENSMUST00000176233.2
|
Gm20695
|
predicted gene 20695 |
| chr16_+_16887991 | 0.34 |
ENSMUST00000232258.2
|
Ypel1
|
yippee like 1 |
| chr2_-_168432235 | 0.33 |
ENSMUST00000109184.8
|
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr10_+_56253418 | 0.32 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr18_+_36693024 | 0.32 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr2_-_148574353 | 0.31 |
ENSMUST00000028926.13
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chrX_+_149330371 | 0.31 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr7_+_24990596 | 0.30 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr17_-_56019571 | 0.29 |
ENSMUST00000086876.7
ENSMUST00000233557.2 ENSMUST00000233972.2 |
Pot1b
|
protection of telomeres 1B |
| chr17_+_44499451 | 0.29 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr5_-_137609691 | 0.28 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr7_-_30672824 | 0.27 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr3_+_37402795 | 0.27 |
ENSMUST00000200585.5
ENSMUST00000038885.10 |
Fgf2
|
fibroblast growth factor 2 |
| chr8_-_103512274 | 0.26 |
ENSMUST00000075190.5
|
Cdh11
|
cadherin 11 |
| chr11_-_106504196 | 0.26 |
ENSMUST00000128933.2
|
Tex2
|
testis expressed gene 2 |
| chr3_-_67282496 | 0.25 |
ENSMUST00000166353.2
|
Gm17402
|
predicted gene, 17402 |
| chr1_-_156767196 | 0.25 |
ENSMUST00000185198.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr19_-_61216834 | 0.23 |
ENSMUST00000076046.7
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr3_-_108747792 | 0.23 |
ENSMUST00000106596.4
ENSMUST00000102621.11 ENSMUST00000138552.6 |
Stxbp3
|
syntaxin binding protein 3 |
| chr17_-_24752683 | 0.23 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr9_-_50650663 | 0.22 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr18_-_46444941 | 0.22 |
ENSMUST00000072835.7
|
Ccdc112
|
coiled-coil domain containing 112 |
| chr1_-_24139263 | 0.21 |
ENSMUST00000187369.7
ENSMUST00000187752.7 ENSMUST00000186999.7 |
Fam135a
|
family with sequence similarity 135, member A |
| chr12_+_4642987 | 0.21 |
ENSMUST00000062580.8
ENSMUST00000220311.2 |
Itsn2
|
intersectin 2 |
| chr5_-_137609634 | 0.21 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr1_-_24139387 | 0.21 |
ENSMUST00000027337.15
|
Fam135a
|
family with sequence similarity 135, member A |
| chr8_-_84874468 | 0.20 |
ENSMUST00000117424.9
ENSMUST00000040383.9 |
Cc2d1a
|
coiled-coil and C2 domain containing 1A |
| chr2_+_112096154 | 0.20 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr6_-_90693471 | 0.20 |
ENSMUST00000101153.10
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr16_+_3702523 | 0.20 |
ENSMUST00000176625.8
ENSMUST00000186375.8 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr1_-_5987617 | 0.19 |
ENSMUST00000044180.5
|
Npbwr1
|
neuropeptides B/W receptor 1 |
| chr12_+_105302853 | 0.19 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr1_-_192880260 | 0.18 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr19_+_24976864 | 0.17 |
ENSMUST00000025831.8
|
Dock8
|
dedicator of cytokinesis 8 |
| chr15_-_77331660 | 0.17 |
ENSMUST00000089469.7
|
Apol7b
|
apolipoprotein L 7b |
| chr5_-_137145030 | 0.16 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr17_-_26063488 | 0.16 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr15_+_77583104 | 0.15 |
ENSMUST00000096358.6
|
Apol7e
|
apolipoprotein L 7e |
| chrX_+_84091915 | 0.15 |
ENSMUST00000239019.2
ENSMUST00000113992.3 ENSMUST00000113991.8 |
Dmd
|
dystrophin, muscular dystrophy |
| chr7_-_65020655 | 0.15 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr1_-_156766957 | 0.14 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr2_+_112097087 | 0.14 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr6_-_53797748 | 0.14 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr15_+_102898966 | 0.13 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chr15_-_71599664 | 0.13 |
ENSMUST00000022953.10
|
Fam135b
|
family with sequence similarity 135, member B |
| chr7_-_30672889 | 0.13 |
ENSMUST00000001279.15
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr9_-_21763898 | 0.13 |
ENSMUST00000217336.2
ENSMUST00000034728.9 |
Dock6
|
dedicator of cytokinesis 6 |
| chr13_-_81781238 | 0.12 |
ENSMUST00000126444.8
ENSMUST00000128585.9 ENSMUST00000146749.2 ENSMUST00000095585.11 |
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr7_+_44465353 | 0.12 |
ENSMUST00000208626.2
ENSMUST00000057195.17 |
Nup62
|
nucleoporin 62 |
| chr10_-_79449826 | 0.11 |
ENSMUST00000059699.9
ENSMUST00000178228.3 |
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chr16_+_42727926 | 0.11 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_156766381 | 0.11 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr1_+_64729950 | 0.11 |
ENSMUST00000187170.7
|
Ccnyl1
|
cyclin Y-like 1 |
| chr8_-_85663976 | 0.11 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr7_-_30672747 | 0.10 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr7_+_100742070 | 0.10 |
ENSMUST00000208439.2
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr11_+_77384234 | 0.10 |
ENSMUST00000037285.10
ENSMUST00000100812.4 |
Git1
|
GIT ArfGAP 1 |
| chr7_+_89779421 | 0.09 |
ENSMUST00000207225.2
ENSMUST00000207484.2 ENSMUST00000209068.2 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr15_+_44320918 | 0.09 |
ENSMUST00000038336.12
ENSMUST00000166957.2 ENSMUST00000209244.2 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr12_+_113061819 | 0.09 |
ENSMUST00000109727.9
ENSMUST00000009099.13 ENSMUST00000109723.8 ENSMUST00000109726.8 ENSMUST00000069690.5 |
Mta1
|
metastasis associated 1 |
| chr11_-_76514334 | 0.09 |
ENSMUST00000238684.2
|
Abr
|
active BCR-related gene |
| chr2_+_121188195 | 0.08 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr11_-_55310724 | 0.08 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr7_-_100504610 | 0.08 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr4_+_40269563 | 0.08 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr7_-_24950145 | 0.08 |
ENSMUST00000116343.3
ENSMUST00000045847.15 |
Erf
|
Ets2 repressor factor |
| chr2_-_164285097 | 0.08 |
ENSMUST00000017153.4
|
Sdc4
|
syndecan 4 |
| chr10_+_116137277 | 0.07 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr3_-_108747767 | 0.07 |
ENSMUST00000196679.5
|
Stxbp3
|
syntaxin binding protein 3 |
| chr6_-_23839136 | 0.07 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr7_+_89779493 | 0.07 |
ENSMUST00000208730.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_+_31750946 | 0.07 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr2_-_30014466 | 0.07 |
ENSMUST00000044751.14
|
Zer1
|
zyg-11 related, cell cycle regulator |
| chr1_+_183078851 | 0.07 |
ENSMUST00000193625.2
|
Aida
|
axin interactor, dorsalization associated |
| chr16_+_16888084 | 0.07 |
ENSMUST00000231514.2
|
Ypel1
|
yippee like 1 |
| chr7_+_104335708 | 0.06 |
ENSMUST00000214876.2
|
Olfr661
|
olfactory receptor 661 |
| chr12_+_4642629 | 0.06 |
ENSMUST00000218402.2
|
Itsn2
|
intersectin 2 |
| chr4_-_129083439 | 0.06 |
ENSMUST00000106059.8
|
S100pbp
|
S100P binding protein |
| chr15_+_34306812 | 0.06 |
ENSMUST00000226766.2
ENSMUST00000163455.9 ENSMUST00000022947.7 ENSMUST00000228570.2 ENSMUST00000227759.2 |
Matn2
|
matrilin 2 |
| chr1_-_183150867 | 0.05 |
ENSMUST00000194543.4
|
Mia3
|
melanoma inhibitory activity 3 |
| chrX_+_151922936 | 0.05 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr12_+_65083093 | 0.05 |
ENSMUST00000120580.8
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr5_+_118307754 | 0.05 |
ENSMUST00000054836.7
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr1_-_155912216 | 0.05 |
ENSMUST00000027738.14
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr19_-_56996617 | 0.05 |
ENSMUST00000118800.8
ENSMUST00000111584.9 ENSMUST00000122359.8 ENSMUST00000148049.8 |
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr14_-_61495934 | 0.05 |
ENSMUST00000077954.13
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr5_-_100521343 | 0.05 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr7_+_89779564 | 0.05 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr6_-_86646118 | 0.05 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr7_+_30673212 | 0.04 |
ENSMUST00000129773.2
|
Fam187b
|
family with sequence similarity 187, member B |
| chr4_-_106474429 | 0.04 |
ENSMUST00000189032.7
ENSMUST00000106788.2 |
Lexm
|
lymphocyte expansion molecule |
| chr16_+_3702604 | 0.04 |
ENSMUST00000115860.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr9_-_27066685 | 0.03 |
ENSMUST00000034472.16
|
Jam3
|
junction adhesion molecule 3 |
| chr7_-_134539993 | 0.03 |
ENSMUST00000171394.3
|
Insyn2a
|
inhibitory synaptic factor 2A |
| chr3_-_57209357 | 0.03 |
ENSMUST00000196979.5
ENSMUST00000029376.13 |
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chrX_+_5959507 | 0.03 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr2_-_169973076 | 0.03 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr17_+_34573760 | 0.02 |
ENSMUST00000178562.2
ENSMUST00000025198.15 |
Btnl2
|
butyrophilin-like 2 |
| chr10_-_79973210 | 0.02 |
ENSMUST00000170219.9
ENSMUST00000169546.9 |
Cbarp
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
| chr13_-_53135064 | 0.02 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr12_-_11200306 | 0.02 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr6_-_85742726 | 0.01 |
ENSMUST00000050780.8
|
Nat8f3
|
N-acetyltransferase 8 (GCN5-related) family member 3 |
| chr1_-_156766351 | 0.01 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr1_+_120530134 | 0.01 |
ENSMUST00000079721.9
|
En1
|
engrailed 1 |
| chr11_+_103540391 | 0.01 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr19_+_4889394 | 0.01 |
ENSMUST00000088653.3
|
Ccdc87
|
coiled-coil domain containing 87 |
| chr2_-_126625343 | 0.01 |
ENSMUST00000028842.9
ENSMUST00000130356.3 |
Usp50
|
ubiquitin specific peptidase 50 |
| chr11_-_107805830 | 0.01 |
ENSMUST00000039071.3
|
Cacng5
|
calcium channel, voltage-dependent, gamma subunit 5 |
| chr10_-_19727165 | 0.01 |
ENSMUST00000059805.6
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr10_+_116013256 | 0.00 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr10_-_109669053 | 0.00 |
ENSMUST00000238286.2
|
Nav3
|
neuron navigator 3 |
| chr2_+_153334710 | 0.00 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr2_+_152753231 | 0.00 |
ENSMUST00000028970.8
|
Mylk2
|
myosin, light polypeptide kinase 2, skeletal muscle |
| chr18_-_82424902 | 0.00 |
ENSMUST00000065224.8
|
Galr1
|
galanin receptor 1 |
| chr7_-_118047394 | 0.00 |
ENSMUST00000203796.4
ENSMUST00000203485.3 |
Syt17
|
synaptotagmin XVII |
| chrX_-_78560891 | 0.00 |
ENSMUST00000197180.6
ENSMUST00000101410.3 |
Cfap47
|
cilia and flagella associated protein 47 |
| chr16_+_20514925 | 0.00 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr7_+_106969919 | 0.00 |
ENSMUST00000137663.2
|
Syt9
|
synaptotagmin IX |
| chrX_-_151552457 | 0.00 |
ENSMUST00000101141.9
ENSMUST00000062317.5 |
Shroom2
|
shroom family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Scrt2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 1.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.3 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.3 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.3 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 0.3 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.5 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.2 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.3 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.0 | 0.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.0 | 0.3 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.3 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 0.3 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.3 | GO:0005922 | fascia adherens(GO:0005916) connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.3 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.2 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 0.3 | GO:1903763 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.5 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |