Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Six3_Six1_Six2
Z-value: 0.18
Transcription factors associated with Six3_Six1_Six2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
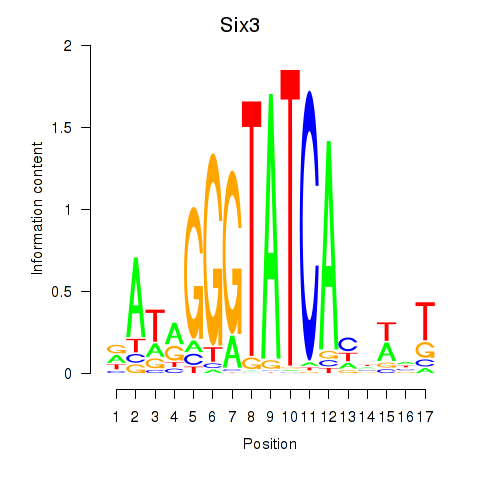
Six3
|
ENSMUSG00000038805.11 | sine oculis-related homeobox 3 |
|
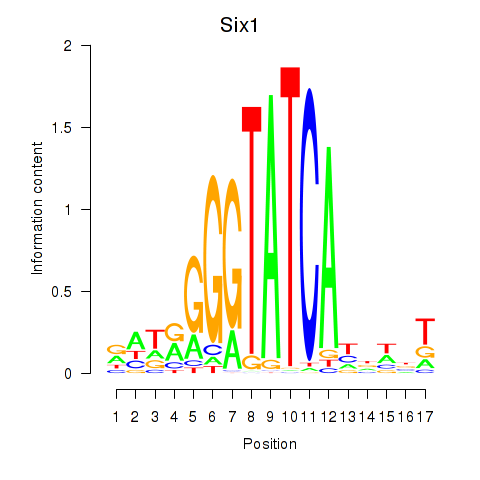
Six1
|
ENSMUSG00000051367.9 | sine oculis-related homeobox 1 |
|
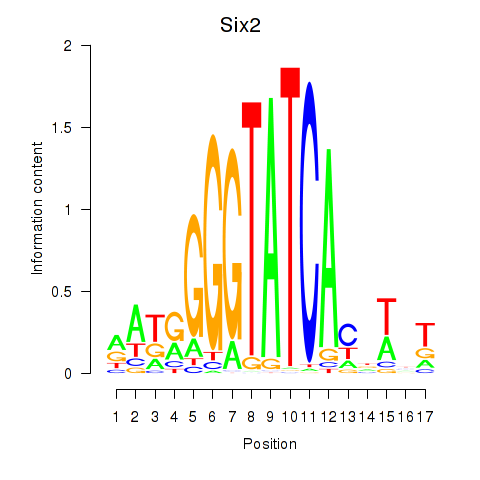
Six2
|
ENSMUSG00000024134.12 | sine oculis-related homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six2 | mm39_v1_chr17_-_85995680_85995703 | 0.97 | 6.2e-03 | Click! |
| Six3 | mm39_v1_chr17_+_85928244_85928270 | 0.93 | 2.0e-02 | Click! |
| Six1 | mm39_v1_chr12_-_73093953_73093953 | 0.04 | 9.5e-01 | Click! |
Activity profile of Six3_Six1_Six2 motif
Sorted Z-values of Six3_Six1_Six2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_126275529 | 0.20 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr9_+_106099797 | 0.18 |
ENSMUST00000062241.11
|
Tlr9
|
toll-like receptor 9 |
| chr7_-_104991477 | 0.14 |
ENSMUST00000213290.2
|
Olfr691
|
olfactory receptor 691 |
| chr2_+_37078210 | 0.13 |
ENSMUST00000213969.2
|
Olfr365
|
olfactory receptor 365 |
| chr2_-_36752671 | 0.13 |
ENSMUST00000213676.2
ENSMUST00000215137.2 |
Olfr351
|
olfactory receptor 351 |
| chr11_+_49379915 | 0.11 |
ENSMUST00000214948.2
|
Olfr1385
|
olfactory receptor 1385 |
| chr7_+_6470100 | 0.10 |
ENSMUST00000207658.4
ENSMUST00000215302.3 |
Olfr1346
|
olfactory receptor 1346 |
| chr6_-_57306479 | 0.09 |
ENSMUST00000227283.2
ENSMUST00000228356.2 |
Vmn1r16
|
vomeronasal 1 receptor 16 |
| chr2_-_88534814 | 0.09 |
ENSMUST00000216928.2
ENSMUST00000216977.2 |
Olfr1196
|
olfactory receptor 1196 |
| chr10_-_81463631 | 0.08 |
ENSMUST00000042923.9
|
Sirt6
|
sirtuin 6 |
| chr12_-_114252202 | 0.08 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr5_+_23992689 | 0.08 |
ENSMUST00000120869.6
ENSMUST00000030852.13 ENSMUST00000117783.8 ENSMUST00000115113.3 |
Rint1
|
RAD50 interactor 1 |
| chr3_+_145926709 | 0.08 |
ENSMUST00000039164.4
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr3_+_94305824 | 0.08 |
ENSMUST00000050975.6
|
Lingo4
|
leucine rich repeat and Ig domain containing 4 |
| chr9_+_38725910 | 0.07 |
ENSMUST00000213164.2
|
Olfr922
|
olfactory receptor 922 |
| chr4_+_132903646 | 0.06 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr7_+_103620359 | 0.06 |
ENSMUST00000209473.4
|
Olfr635
|
olfactory receptor 635 |
| chr9_-_70411000 | 0.06 |
ENSMUST00000034739.12
|
Rnf111
|
ring finger 111 |
| chr5_+_139408906 | 0.05 |
ENSMUST00000066211.5
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr14_+_58035640 | 0.05 |
ENSMUST00000111269.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr9_+_98305014 | 0.05 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chr15_-_76090508 | 0.04 |
ENSMUST00000073418.13
ENSMUST00000171634.8 ENSMUST00000076442.12 |
Plec
|
plectin |
| chr7_-_102507962 | 0.04 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr10_-_12744025 | 0.04 |
ENSMUST00000219660.2
|
Utrn
|
utrophin |
| chr2_-_89408791 | 0.04 |
ENSMUST00000217402.2
|
Olfr1245
|
olfactory receptor 1245 |
| chr10_-_12743915 | 0.04 |
ENSMUST00000219584.2
|
Utrn
|
utrophin |
| chr16_-_18052937 | 0.04 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr19_+_55304703 | 0.03 |
ENSMUST00000225529.2
ENSMUST00000223690.2 ENSMUST00000095950.3 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr5_+_21748523 | 0.03 |
ENSMUST00000035651.6
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr17_-_53846438 | 0.03 |
ENSMUST00000056198.4
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr7_+_102965522 | 0.03 |
ENSMUST00000216456.2
|
Olfr597
|
olfactory receptor 597 |
| chr15_-_66985760 | 0.02 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr17_+_35460722 | 0.02 |
ENSMUST00000068056.12
ENSMUST00000174757.8 ENSMUST00000173731.8 |
Ddx39b
|
DEAD box helicase 39b |
| chr11_+_117688486 | 0.02 |
ENSMUST00000106331.2
|
6030468B19Rik
|
RIKEN cDNA 6030468B19 gene |
| chr9_+_123921573 | 0.02 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr6_+_123100272 | 0.02 |
ENSMUST00000041779.13
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr9_+_103879745 | 0.02 |
ENSMUST00000035167.15
ENSMUST00000194774.6 |
Nphp3
|
nephronophthisis 3 (adolescent) |
| chr1_+_139382485 | 0.02 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr2_+_157870399 | 0.02 |
ENSMUST00000103123.10
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr2_-_13276074 | 0.02 |
ENSMUST00000137670.3
ENSMUST00000114791.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr11_+_120612278 | 0.02 |
ENSMUST00000018156.12
|
Rac3
|
Rac family small GTPase 3 |
| chr8_+_72704882 | 0.02 |
ENSMUST00000131237.8
ENSMUST00000136516.9 ENSMUST00000109997.10 ENSMUST00000132848.2 |
Zfp961
|
zinc finger protein 961 |
| chr8_-_87611849 | 0.02 |
ENSMUST00000034074.8
|
N4bp1
|
NEDD4 binding protein 1 |
| chr2_-_13276205 | 0.02 |
ENSMUST00000191959.6
ENSMUST00000028059.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr17_+_41121979 | 0.02 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr6_+_123100382 | 0.02 |
ENSMUST00000032248.8
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr7_+_43339842 | 0.02 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr14_+_58035614 | 0.02 |
ENSMUST00000128764.9
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr11_-_101877832 | 0.02 |
ENSMUST00000107173.9
ENSMUST00000107172.8 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr6_-_119925387 | 0.02 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr17_+_24026892 | 0.02 |
ENSMUST00000191385.3
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr10_-_128016135 | 0.01 |
ENSMUST00000238843.2
ENSMUST00000099139.9 |
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr8_-_23143422 | 0.01 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta |
| chr7_-_102368277 | 0.01 |
ENSMUST00000216312.2
|
Olfr33
|
olfactory receptor 33 |
| chr7_+_29931309 | 0.01 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr9_+_72866067 | 0.01 |
ENSMUST00000098567.9
ENSMUST00000034734.9 |
Dnaaf4
|
dynein axonemal assembly factor 4 |
| chr11_-_73800125 | 0.01 |
ENSMUST00000215690.2
|
Olfr395
|
olfactory receptor 395 |
| chr7_-_100504610 | 0.01 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr16_-_45544960 | 0.01 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr9_-_39920878 | 0.01 |
ENSMUST00000216463.3
|
Olfr980
|
olfactory receptor 980 |
| chr15_-_75766321 | 0.01 |
ENSMUST00000023237.8
|
Naprt
|
nicotinate phosphoribosyltransferase |
| chr17_+_36176485 | 0.01 |
ENSMUST00000127442.8
ENSMUST00000144382.8 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_+_136395673 | 0.01 |
ENSMUST00000189413.7
ENSMUST00000047817.12 |
Kif14
|
kinesin family member 14 |
| chr15_+_82136598 | 0.01 |
ENSMUST00000136948.3
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr11_+_120612369 | 0.01 |
ENSMUST00000142229.2
|
Rac3
|
Rac family small GTPase 3 |
| chr9_-_119170456 | 0.01 |
ENSMUST00000139870.2
|
Myd88
|
myeloid differentiation primary response gene 88 |
| chr17_-_56312555 | 0.00 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr12_-_114451189 | 0.00 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr16_-_31094095 | 0.00 |
ENSMUST00000060188.14
|
Ppp1r2
|
protein phosphatase 1, regulatory inhibitor subunit 2 |
| chr15_+_101152078 | 0.00 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr7_+_15484302 | 0.00 |
ENSMUST00000098802.10
ENSMUST00000173053.2 |
Obox5
|
oocyte specific homeobox 5 |
| chr6_-_69741999 | 0.00 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr18_+_89224219 | 0.00 |
ENSMUST00000236835.2
|
Cd226
|
CD226 antigen |
| chr7_-_103393740 | 0.00 |
ENSMUST00000216300.2
|
Olfr629
|
olfactory receptor 629 |
| chr7_-_48149069 | 0.00 |
ENSMUST00000098433.5
|
Mrgprx2
|
MAS-related GPR, member X2 |
| chr14_-_75830550 | 0.00 |
ENSMUST00000164082.9
ENSMUST00000169658.9 |
Cby2
|
chibby family member 2 |
| chr8_+_70285282 | 0.00 |
ENSMUST00000131637.9
|
Pbx4
|
pre B cell leukemia homeobox 4 |
| chr9_-_121668527 | 0.00 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr11_-_6180127 | 0.00 |
ENSMUST00000004505.3
|
Npc1l1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr7_+_43837629 | 0.00 |
ENSMUST00000073713.8
|
Klk1b24
|
kallikrein 1-related peptidase b24 |
| chrY_-_13223928 | 0.00 |
ENSMUST00000178050.2
|
Gm21828
|
predicted gene, 21828 |
| chrY_-_13349063 | 0.00 |
ENSMUST00000178519.2
|
Gm21725
|
predicted gene, 21725 |
| chrY_-_18061812 | 0.00 |
ENSMUST00000179219.2
|
Gm21764
|
predicted gene, 21764 |
| chr5_+_138805835 | 0.00 |
ENSMUST00000179205.2
|
Foxl3
|
forkhead box L3 |
| chr16_-_58647137 | 0.00 |
ENSMUST00000215069.2
ENSMUST00000079955.6 |
Olfr175
|
olfactory receptor 175 |
| chr14_+_53791444 | 0.00 |
ENSMUST00000198297.2
|
Trav14-1
|
T cell receptor alpha variable 14-1 |
| chr2_-_76698725 | 0.00 |
ENSMUST00000149616.8
ENSMUST00000152185.8 ENSMUST00000130915.8 ENSMUST00000155365.8 ENSMUST00000128071.8 |
Ttn
|
titin |
| chr3_-_88332401 | 0.00 |
ENSMUST00000168755.7
ENSMUST00000193433.6 ENSMUST00000195657.6 ENSMUST00000057935.9 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr11_-_69586347 | 0.00 |
ENSMUST00000181261.2
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chrY_-_40731110 | 0.00 |
ENSMUST00000177579.2
|
Gm21797
|
predicted gene, 21797 |
| chr6_+_48514518 | 0.00 |
ENSMUST00000040361.8
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2 |
| chr6_+_48514578 | 0.00 |
ENSMUST00000203011.2
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2 |
| chrY_+_13941738 | 0.00 |
ENSMUST00000178689.2
|
Gm21767
|
predicted gene, 21767 |
| chrY_+_18654482 | 0.00 |
ENSMUST00000179302.2
|
Gm21822
|
predicted gene, 21822 |
| chr1_-_83020201 | 0.00 |
ENSMUST00000188323.2
|
Krtap28-10
|
keratin associated protein 28-10 |
| chrY_-_13465579 | 0.00 |
ENSMUST00000178428.2
|
Gm21866
|
predicted gene, 21866 |
| chr6_+_38639945 | 0.00 |
ENSMUST00000114874.5
|
Clec2l
|
C-type lectin domain family 2, member L |
Network of associatons between targets according to the STRING database.
First level regulatory network of Six3_Six1_Six2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:2000722 | response to mineralocorticoid(GO:0051385) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |