Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
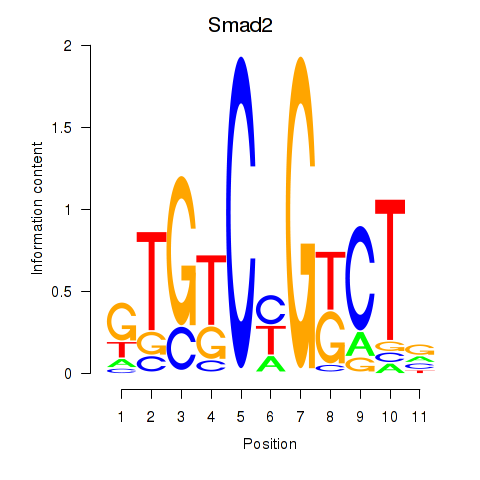
Results for Smad2
Z-value: 0.56
Transcription factors associated with Smad2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad2
|
ENSMUSG00000024563.17 | SMAD family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad2 | mm39_v1_chr18_+_76374959_76375117 | -0.85 | 6.5e-02 | Click! |
Activity profile of Smad2 motif
Sorted Z-values of Smad2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23936250 | 1.03 |
ENSMUST00000091703.3
|
H3c2
|
H3 clustered histone 2 |
| chr2_-_143853122 | 0.45 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr10_-_49664839 | 0.35 |
ENSMUST00000220263.2
ENSMUST00000218823.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr12_-_113271532 | 0.29 |
ENSMUST00000192188.3
ENSMUST00000103418.3 |
Ighg2b
|
immunoglobulin heavy constant gamma 2B |
| chr4_+_83335947 | 0.29 |
ENSMUST00000030206.10
ENSMUST00000071544.11 |
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr7_+_142030744 | 0.28 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr6_-_124733441 | 0.26 |
ENSMUST00000088357.12
|
Atn1
|
atrophin 1 |
| chr12_+_33197776 | 0.26 |
ENSMUST00000125192.9
ENSMUST00000136644.8 |
Atxn7l1
|
ataxin 7-like 1 |
| chr8_+_91635192 | 0.24 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr12_+_33197753 | 0.23 |
ENSMUST00000146040.9
|
Atxn7l1
|
ataxin 7-like 1 |
| chr3_+_135143910 | 0.23 |
ENSMUST00000196446.5
ENSMUST00000106291.10 ENSMUST00000199613.5 |
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr13_-_100240570 | 0.22 |
ENSMUST00000038104.12
|
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr16_-_17540805 | 0.21 |
ENSMUST00000012259.9
ENSMUST00000232236.2 |
Med15
|
mediator complex subunit 15 |
| chr3_+_135144304 | 0.19 |
ENSMUST00000198685.5
ENSMUST00000197859.5 |
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_-_113294138 | 0.19 |
ENSMUST00000194304.6
ENSMUST00000103420.3 |
Ighg1
|
immunoglobulin heavy constant gamma 1 (G1m marker) |
| chr3_-_75864195 | 0.17 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr8_+_66838927 | 0.15 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr9_+_110074574 | 0.13 |
ENSMUST00000197850.5
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr6_-_113354826 | 0.13 |
ENSMUST00000032410.14
|
Tada3
|
transcriptional adaptor 3 |
| chr1_-_160079007 | 0.12 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr6_+_54017063 | 0.12 |
ENSMUST00000127323.3
|
Chn2
|
chimerin 2 |
| chr3_+_135144202 | 0.10 |
ENSMUST00000166033.6
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr9_-_65487322 | 0.10 |
ENSMUST00000068944.9
|
Plekho2
|
pleckstrin homology domain containing, family O member 2 |
| chr17_-_24752683 | 0.10 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr8_-_106363982 | 0.09 |
ENSMUST00000182863.2
ENSMUST00000194091.6 |
Gm5914
Agrp
|
predicted gene 5914 agouti related neuropeptide |
| chr4_+_42950367 | 0.09 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr6_-_90758954 | 0.09 |
ENSMUST00000238821.2
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr7_-_44702269 | 0.08 |
ENSMUST00000057293.8
|
Prr12
|
proline rich 12 |
| chr9_+_123307758 | 0.08 |
ENSMUST00000026269.4
|
Limd1
|
LIM domains containing 1 |
| chr3_+_135144287 | 0.08 |
ENSMUST00000196591.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr16_+_18166045 | 0.08 |
ENSMUST00000239533.1
ENSMUST00000239534.1 |
ARVCF
|
armadillo repeat deleted in velocardiofacial syndrome |
| chr6_+_54016543 | 0.07 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr6_-_142647944 | 0.07 |
ENSMUST00000100827.5
ENSMUST00000087527.11 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr14_-_20546848 | 0.06 |
ENSMUST00000022353.5
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr17_+_36176485 | 0.06 |
ENSMUST00000127442.8
ENSMUST00000144382.8 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr17_+_36176948 | 0.05 |
ENSMUST00000122899.8
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr2_-_148285450 | 0.05 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr2_+_38229270 | 0.05 |
ENSMUST00000143783.9
|
Lhx2
|
LIM homeobox protein 2 |
| chr2_-_34262012 | 0.05 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr4_+_11191354 | 0.04 |
ENSMUST00000170901.8
|
Ccne2
|
cyclin E2 |
| chr14_+_4137837 | 0.04 |
ENSMUST00000022296.7
|
Ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr5_-_146521629 | 0.04 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr17_-_34834938 | 0.04 |
ENSMUST00000097345.10
ENSMUST00000015611.14 |
Egfl8
|
EGF-like domain 8 |
| chr6_-_142647985 | 0.04 |
ENSMUST00000205202.3
ENSMUST00000073173.12 ENSMUST00000111771.8 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr7_-_126062272 | 0.03 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr15_-_84007373 | 0.03 |
ENSMUST00000230566.2
ENSMUST00000019012.4 |
Pnpla5
|
patatin-like phospholipase domain containing 5 |
| chr15_+_82136598 | 0.02 |
ENSMUST00000136948.3
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr4_+_117706390 | 0.02 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr18_-_42712717 | 0.02 |
ENSMUST00000054738.5
|
Gpr151
|
G protein-coupled receptor 151 |
| chr12_+_51640097 | 0.02 |
ENSMUST00000164782.10
ENSMUST00000085412.7 |
Coch
|
cochlin |
| chr17_+_3447465 | 0.02 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chrX_-_20797736 | 0.02 |
ENSMUST00000001156.8
|
Cfp
|
complement factor properdin |
| chr1_-_150868802 | 0.01 |
ENSMUST00000074783.12
ENSMUST00000137197.9 |
Hmcn1
|
hemicentin 1 |
| chr16_-_34334314 | 0.01 |
ENSMUST00000151491.8
ENSMUST00000114960.9 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr4_+_117692583 | 0.01 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr19_+_5928649 | 0.01 |
ENSMUST00000136833.8
ENSMUST00000141362.2 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr3_+_135144481 | 0.01 |
ENSMUST00000199582.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_30639217 | 0.01 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr4_-_126056412 | 0.00 |
ENSMUST00000094760.9
|
Sh3d21
|
SH3 domain containing 21 |
| chr16_-_34334454 | 0.00 |
ENSMUST00000089655.12
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr4_+_115741581 | 0.00 |
ENSMUST00000097918.3
|
Kncn
|
kinocilin |
| chr9_+_101815680 | 0.00 |
ENSMUST00000112841.2
|
9630041A04Rik
|
RIKEN cDNA 9630041A04 gene |
| chr11_+_63022328 | 0.00 |
ENSMUST00000018361.10
|
Pmp22
|
peripheral myelin protein 22 |
| chr4_-_126057263 | 0.00 |
ENSMUST00000097891.4
|
Sh3d21
|
SH3 domain containing 21 |
| chr1_-_173195236 | 0.00 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr10_-_77533067 | 0.00 |
ENSMUST00000220084.2
|
Gm9508
|
predicted gene 9508 |
| chr11_+_69737437 | 0.00 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr17_-_34834745 | 0.00 |
ENSMUST00000168353.4
|
Egfl8
|
EGF-like domain 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.3 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0090076 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |