Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
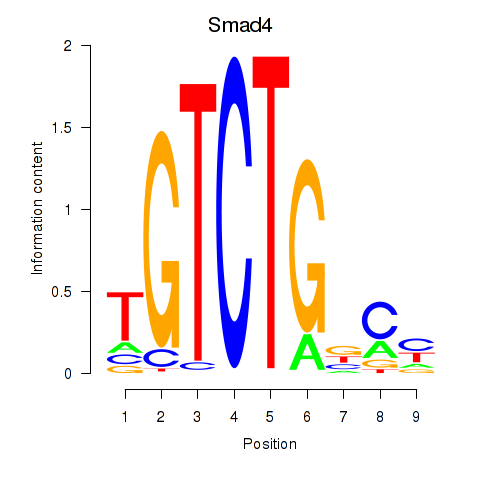
Results for Smad4
Z-value: 3.17
Transcription factors associated with Smad4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad4
|
ENSMUSG00000024515.14 | SMAD family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad4 | mm39_v1_chr18_-_73836810_73836877 | 0.86 | 6.2e-02 | Click! |
Activity profile of Smad4 motif
Sorted Z-values of Smad4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23765546 | 5.96 |
ENSMUST00000102968.3
|
H4c4
|
H4 clustered histone 4 |
| chr13_+_22227359 | 5.93 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr13_+_23715220 | 5.55 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr13_-_23735822 | 5.42 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr3_-_96128196 | 5.15 |
ENSMUST00000090782.4
|
H2ac20
|
H2A clustered histone 20 |
| chr13_-_22016364 | 5.02 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr13_+_21995906 | 4.53 |
ENSMUST00000104941.4
|
H4c17
|
H4 clustered histone 17 |
| chr15_+_102885467 | 3.65 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9 |
| chrX_-_71318353 | 3.64 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr13_-_24118139 | 2.86 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr13_-_22227114 | 2.72 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr11_-_69304501 | 2.43 |
ENSMUST00000094077.5
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr1_-_74130488 | 2.35 |
ENSMUST00000187584.7
|
Tns1
|
tensin 1 |
| chr2_-_121211410 | 1.83 |
ENSMUST00000038389.15
|
Strc
|
stereocilin |
| chr7_+_119495058 | 1.59 |
ENSMUST00000106518.9
ENSMUST00000207270.2 ENSMUST00000208424.2 ENSMUST00000208202.2 ENSMUST00000054440.11 |
Lyrm1
|
LYR motif containing 1 |
| chr15_+_6599001 | 1.54 |
ENSMUST00000227175.2
|
Fyb
|
FYN binding protein |
| chr9_-_49710058 | 1.46 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr5_+_23639432 | 1.43 |
ENSMUST00000094962.9
ENSMUST00000115128.8 |
Kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr11_+_105866030 | 1.35 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr19_+_4282487 | 1.27 |
ENSMUST00000235306.2
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chrX_+_158491589 | 1.21 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr9_-_49710190 | 1.19 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr1_+_59802543 | 1.17 |
ENSMUST00000087435.7
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr1_+_176642226 | 1.16 |
ENSMUST00000056773.15
ENSMUST00000027785.15 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr1_-_176641607 | 1.16 |
ENSMUST00000195717.6
ENSMUST00000192961.6 |
Cep170
|
centrosomal protein 170 |
| chr19_+_4147391 | 1.12 |
ENSMUST00000174514.2
ENSMUST00000174149.8 |
Cdk2ap2
|
CDK2-associated protein 2 |
| chr16_+_43067641 | 1.11 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_-_102871440 | 1.11 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr6_-_149003003 | 1.10 |
ENSMUST00000127727.2
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr4_-_119047167 | 1.10 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_119047202 | 1.07 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_148215135 | 1.02 |
ENSMUST00000030862.5
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr13_-_42000958 | 0.98 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr15_-_103231921 | 0.91 |
ENSMUST00000229551.2
|
Zfp385a
|
zinc finger protein 385A |
| chr19_-_40260060 | 0.89 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr3_+_63203235 | 0.87 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr7_+_25386418 | 0.82 |
ENSMUST00000002678.10
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr2_+_152578164 | 0.81 |
ENSMUST00000038368.9
ENSMUST00000109824.2 |
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chr2_+_29759495 | 0.79 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chrX_-_7537580 | 0.79 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr19_+_4281953 | 0.78 |
ENSMUST00000025773.5
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr2_-_45001141 | 0.78 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr16_-_44978546 | 0.77 |
ENSMUST00000114600.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr11_-_4110286 | 0.76 |
ENSMUST00000093381.11
ENSMUST00000101626.9 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr4_+_150171822 | 0.72 |
ENSMUST00000094451.4
|
Gpr157
|
G protein-coupled receptor 157 |
| chr4_-_148584906 | 0.69 |
ENSMUST00000030840.4
|
Angptl7
|
angiopoietin-like 7 |
| chr15_-_103161237 | 0.68 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr2_+_111205867 | 0.68 |
ENSMUST00000062407.6
|
Olfr1284
|
olfactory receptor 1284 |
| chr4_+_42950367 | 0.62 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr7_+_101010447 | 0.61 |
ENSMUST00000137384.8
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr14_+_76652369 | 0.61 |
ENSMUST00000110888.8
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr2_-_155676765 | 0.60 |
ENSMUST00000029143.7
ENSMUST00000239423.2 |
Fam83c
|
family with sequence similarity 83, member C |
| chr17_-_30107544 | 0.60 |
ENSMUST00000171691.9
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr6_+_54016543 | 0.59 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr9_+_61280501 | 0.58 |
ENSMUST00000162583.8
ENSMUST00000161993.8 ENSMUST00000160882.8 ENSMUST00000160724.8 ENSMUST00000162973.8 ENSMUST00000159050.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr7_+_127084283 | 0.58 |
ENSMUST00000048896.8
|
Fbrs
|
fibrosin |
| chr9_+_61280764 | 0.57 |
ENSMUST00000160541.8
ENSMUST00000161207.8 ENSMUST00000159630.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr11_+_59433554 | 0.57 |
ENSMUST00000149126.2
|
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr12_+_36431449 | 0.53 |
ENSMUST00000221452.2
ENSMUST00000062041.6 ENSMUST00000220519.2 ENSMUST00000221895.2 |
Crppa
|
CDP-L-ribitol pyrophosphorylase A |
| chr6_-_90758954 | 0.52 |
ENSMUST00000238821.2
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr3_-_96833336 | 0.51 |
ENSMUST00000062944.7
|
Gja8
|
gap junction protein, alpha 8 |
| chr7_+_55417967 | 0.51 |
ENSMUST00000060416.15
ENSMUST00000094360.13 ENSMUST00000165045.9 ENSMUST00000173835.2 |
Siglech
|
sialic acid binding Ig-like lectin H |
| chrX_+_150586309 | 0.50 |
ENSMUST00000026292.15
|
Huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr7_+_44865177 | 0.49 |
ENSMUST00000033060.14
ENSMUST00000210447.2 ENSMUST00000211744.2 ENSMUST00000107801.10 |
Tead2
|
TEA domain family member 2 |
| chr14_+_65187485 | 0.49 |
ENSMUST00000043914.8
ENSMUST00000239450.2 |
Ints9
|
integrator complex subunit 9 |
| chr9_+_61280890 | 0.48 |
ENSMUST00000161689.8
|
Tle3
|
transducin-like enhancer of split 3 |
| chr11_-_106050927 | 0.47 |
ENSMUST00000045923.10
|
Limd2
|
LIM domain containing 2 |
| chr5_-_137739364 | 0.47 |
ENSMUST00000149512.3
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chrX_-_37653396 | 0.46 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chr5_+_64960705 | 0.45 |
ENSMUST00000165536.8
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr5_+_120569764 | 0.43 |
ENSMUST00000031591.10
|
Lhx5
|
LIM homeobox protein 5 |
| chr2_+_96148418 | 0.42 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr11_-_5394838 | 0.42 |
ENSMUST00000109867.8
ENSMUST00000143746.3 |
Znrf3
|
zinc and ring finger 3 |
| chr6_+_54017063 | 0.41 |
ENSMUST00000127323.3
|
Chn2
|
chimerin 2 |
| chr11_+_4110346 | 0.40 |
ENSMUST00000002198.4
|
Sf3a1
|
splicing factor 3a, subunit 1 |
| chr15_-_96358612 | 0.39 |
ENSMUST00000047835.8
|
Scaf11
|
SR-related CTD-associated factor 11 |
| chr6_-_129428869 | 0.38 |
ENSMUST00000203162.3
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr1_+_63655127 | 0.38 |
ENSMUST00000226288.2
|
Gm39653
|
predicted gene, 39653 |
| chr1_-_133728779 | 0.36 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr11_-_3454766 | 0.36 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr19_-_40260286 | 0.36 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr14_-_65187076 | 0.36 |
ENSMUST00000176832.8
|
Hmbox1
|
homeobox containing 1 |
| chr15_-_38299934 | 0.36 |
ENSMUST00000228772.2
|
Klf10
|
Kruppel-like factor 10 |
| chr4_+_97660971 | 0.36 |
ENSMUST00000152023.8
|
Nfia
|
nuclear factor I/A |
| chr15_-_38300937 | 0.35 |
ENSMUST00000227920.2
ENSMUST00000074043.7 ENSMUST00000228416.2 |
Klf10
|
Kruppel-like factor 10 |
| chr14_-_65187287 | 0.35 |
ENSMUST00000067843.10
ENSMUST00000176489.8 ENSMUST00000175905.8 ENSMUST00000022544.14 ENSMUST00000175744.8 ENSMUST00000176128.8 |
Hmbox1
|
homeobox containing 1 |
| chr19_+_5790918 | 0.35 |
ENSMUST00000081496.6
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr15_+_81820954 | 0.35 |
ENSMUST00000038757.8
ENSMUST00000230633.2 |
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr17_-_35381945 | 0.34 |
ENSMUST00000174805.2
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr14_+_4137837 | 0.34 |
ENSMUST00000022296.7
|
Ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr7_-_4781140 | 0.33 |
ENSMUST00000094892.12
|
Il11
|
interleukin 11 |
| chr13_+_24685508 | 0.33 |
ENSMUST00000238974.2
|
Ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr1_-_64160557 | 0.32 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr10_+_33780993 | 0.30 |
ENSMUST00000169670.8
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr5_+_65694687 | 0.30 |
ENSMUST00000200946.3
ENSMUST00000201383.2 |
Ube2k
|
ubiquitin-conjugating enzyme E2K |
| chr5_+_65694598 | 0.29 |
ENSMUST00000201266.4
ENSMUST00000201292.4 ENSMUST00000201984.4 ENSMUST00000202679.4 ENSMUST00000202082.4 |
Ube2k
|
ubiquitin-conjugating enzyme E2K |
| chr7_+_116980173 | 0.29 |
ENSMUST00000032892.7
|
Xylt1
|
xylosyltransferase 1 |
| chr5_+_111565129 | 0.28 |
ENSMUST00000094463.5
|
Mn1
|
meningioma 1 |
| chr5_+_117979899 | 0.28 |
ENSMUST00000142742.9
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr17_-_47169380 | 0.28 |
ENSMUST00000233455.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr10_+_79986988 | 0.28 |
ENSMUST00000146516.8
ENSMUST00000144526.2 |
Midn
|
midnolin |
| chr11_-_3881995 | 0.27 |
ENSMUST00000020710.11
ENSMUST00000109989.10 ENSMUST00000109991.8 ENSMUST00000109993.9 |
Tcn2
|
transcobalamin 2 |
| chr4_+_132738787 | 0.26 |
ENSMUST00000105915.8
ENSMUST00000105916.8 |
Ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr18_-_33596890 | 0.26 |
ENSMUST00000237066.2
|
Nrep
|
neuronal regeneration related protein |
| chr5_+_65694552 | 0.24 |
ENSMUST00000142407.8
|
Ube2k
|
ubiquitin-conjugating enzyme E2K |
| chr17_+_38082190 | 0.24 |
ENSMUST00000217119.2
|
Olfr122
|
olfactory receptor 122 |
| chr11_-_3881789 | 0.23 |
ENSMUST00000109992.8
ENSMUST00000109988.2 |
Tcn2
|
transcobalamin 2 |
| chr11_+_11414256 | 0.23 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr11_-_118233326 | 0.23 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr11_+_101875095 | 0.22 |
ENSMUST00000176722.8
ENSMUST00000175972.2 |
Cfap97d1
|
CFAP97 domain containing 1 |
| chr11_-_3881960 | 0.22 |
ENSMUST00000109990.8
|
Tcn2
|
transcobalamin 2 |
| chr16_+_34605282 | 0.21 |
ENSMUST00000023538.9
|
Mylk
|
myosin, light polypeptide kinase |
| chr6_+_121320008 | 0.21 |
ENSMUST00000166457.8
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr2_+_70393782 | 0.20 |
ENSMUST00000123330.3
|
Gad1
|
glutamate decarboxylase 1 |
| chr19_-_10716335 | 0.20 |
ENSMUST00000025571.9
ENSMUST00000238167.2 |
Cd5
|
CD5 antigen |
| chr12_-_109566764 | 0.19 |
ENSMUST00000149046.4
|
Rtl1
|
retrotransposon Gaglike 1 |
| chr15_+_82230155 | 0.19 |
ENSMUST00000023086.15
|
Smdt1
|
single-pass membrane protein with aspartate rich tail 1 |
| chr6_+_38410848 | 0.18 |
ENSMUST00000160583.8
|
Ubn2
|
ubinuclein 2 |
| chrX_-_149371067 | 0.18 |
ENSMUST00000140207.8
ENSMUST00000112719.2 ENSMUST00000112727.10 ENSMUST00000112721.10 ENSMUST00000026303.16 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr6_-_30693675 | 0.18 |
ENSMUST00000169422.8
ENSMUST00000115131.8 ENSMUST00000115130.3 ENSMUST00000031810.15 |
Cep41
|
centrosomal protein 41 |
| chr11_-_101062111 | 0.18 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr14_-_30890544 | 0.18 |
ENSMUST00000036618.14
|
Stab1
|
stabilin 1 |
| chr14_-_25903524 | 0.18 |
ENSMUST00000052286.16
|
Plac9a
|
placenta specific 9a |
| chr14_-_20546848 | 0.18 |
ENSMUST00000022353.5
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr11_+_53324126 | 0.17 |
ENSMUST00000018382.7
|
Gdf9
|
growth differentiation factor 9 |
| chr16_+_43056218 | 0.17 |
ENSMUST00000146708.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr11_+_69737437 | 0.17 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr7_-_131918926 | 0.17 |
ENSMUST00000080215.6
|
Chst15
|
carbohydrate sulfotransferase 15 |
| chr7_-_70010341 | 0.16 |
ENSMUST00000032768.15
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_68414401 | 0.15 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr1_-_57010921 | 0.15 |
ENSMUST00000114415.10
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr9_+_21336300 | 0.15 |
ENSMUST00000172482.8
ENSMUST00000174050.8 |
Dnm2
|
dynamin 2 |
| chr7_-_119494669 | 0.15 |
ENSMUST00000098080.9
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr11_-_103099349 | 0.15 |
ENSMUST00000174567.3
ENSMUST00000021323.11 ENSMUST00000107026.9 |
Efcab15
|
EF-hand calcium binding domain 15 |
| chr13_-_42001102 | 0.14 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr4_+_130640611 | 0.14 |
ENSMUST00000156225.8
ENSMUST00000156742.8 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr18_-_33596792 | 0.14 |
ENSMUST00000051087.16
|
Nrep
|
neuronal regeneration related protein |
| chr7_-_119494918 | 0.14 |
ENSMUST00000059851.14
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr3_-_135397298 | 0.13 |
ENSMUST00000029812.14
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr11_-_115310743 | 0.12 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr6_+_125122172 | 0.12 |
ENSMUST00000119527.8
ENSMUST00000117675.8 |
Iffo1
|
intermediate filament family orphan 1 |
| chr7_+_4693759 | 0.11 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr17_+_75485791 | 0.11 |
ENSMUST00000135447.8
ENSMUST00000112516.8 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_31530049 | 0.11 |
ENSMUST00000113470.3
|
Prdm12
|
PR domain containing 12 |
| chr17_-_25300112 | 0.11 |
ENSMUST00000024984.7
|
Tmem204
|
transmembrane protein 204 |
| chr3_+_60436570 | 0.11 |
ENSMUST00000192607.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chrX_-_7765171 | 0.10 |
ENSMUST00000115655.8
ENSMUST00000156741.8 |
Pqbp1
|
polyglutamine binding protein 1 |
| chr17_+_34482183 | 0.10 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr17_+_34372046 | 0.10 |
ENSMUST00000114232.4
|
H2-DMb1
|
histocompatibility 2, class II, locus Mb1 |
| chr7_-_126496534 | 0.09 |
ENSMUST00000120007.8
|
Tmem219
|
transmembrane protein 219 |
| chr7_+_45434755 | 0.09 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr2_+_125876566 | 0.09 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr8_-_41469786 | 0.08 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr12_-_112766266 | 0.08 |
ENSMUST00000239525.1
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr17_-_90395771 | 0.08 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr4_+_127881786 | 0.08 |
ENSMUST00000184063.3
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr14_-_55762432 | 0.07 |
ENSMUST00000062232.15
|
Nrl
|
neural retina leucine zipper gene |
| chr3_-_116762617 | 0.07 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr15_+_102012782 | 0.07 |
ENSMUST00000230474.2
|
Tns2
|
tensin 2 |
| chr14_-_55762416 | 0.07 |
ENSMUST00000178694.3
|
Nrl
|
neural retina leucine zipper gene |
| chr11_+_95715295 | 0.07 |
ENSMUST00000150134.2
ENSMUST00000054173.4 |
Phospho1
|
phosphatase, orphan 1 |
| chr7_-_30062197 | 0.07 |
ENSMUST00000046351.7
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr10_-_127370408 | 0.07 |
ENSMUST00000095266.3
|
Nxph4
|
neurexophilin 4 |
| chr7_+_143729250 | 0.07 |
ENSMUST00000105900.9
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr9_-_58065800 | 0.06 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr8_-_49008305 | 0.06 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr1_-_74627264 | 0.06 |
ENSMUST00000066986.13
|
Zfp142
|
zinc finger protein 142 |
| chr11_+_115331365 | 0.06 |
ENSMUST00000093914.5
|
Trim80
|
tripartite motif-containing 80 |
| chr6_+_113448388 | 0.06 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr2_+_125876883 | 0.06 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr12_+_113453131 | 0.06 |
ENSMUST00000063317.4
|
Adam6b
|
a disintegrin and metallopeptidase domain 6B |
| chr15_+_89417017 | 0.06 |
ENSMUST00000167173.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr2_+_70393195 | 0.06 |
ENSMUST00000130998.8
|
Gad1
|
glutamate decarboxylase 1 |
| chr11_+_59995796 | 0.06 |
ENSMUST00000064190.13
|
Rai1
|
retinoic acid induced 1 |
| chr8_+_96078886 | 0.06 |
ENSMUST00000034243.7
|
Mmp15
|
matrix metallopeptidase 15 |
| chr3_-_116762476 | 0.06 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr7_-_45519853 | 0.05 |
ENSMUST00000211713.2
|
Grin2d
|
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
| chr9_+_122717536 | 0.05 |
ENSMUST00000063980.8
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr2_+_31777273 | 0.05 |
ENSMUST00000138325.8
ENSMUST00000028187.7 |
Lamc3
|
laminin gamma 3 |
| chr18_+_60659257 | 0.04 |
ENSMUST00000223984.2
ENSMUST00000025505.7 ENSMUST00000223590.2 |
Dctn4
|
dynactin 4 |
| chr7_+_6572992 | 0.04 |
ENSMUST00000070985.4
|
Olfr1350
|
olfactory receptor 1350 |
| chr15_+_102011352 | 0.04 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr5_-_41921834 | 0.04 |
ENSMUST00000060820.8
|
Nkx3-2
|
NK3 homeobox 2 |
| chr8_+_79754980 | 0.04 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr18_+_69479211 | 0.03 |
ENSMUST00000201235.4
|
Tcf4
|
transcription factor 4 |
| chr17_+_75485906 | 0.03 |
ENSMUST00000112514.2
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr5_+_73164226 | 0.03 |
ENSMUST00000031127.11
ENSMUST00000201304.2 |
Slc10a4
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 4 |
| chr1_+_162466717 | 0.03 |
ENSMUST00000028020.11
|
Myoc
|
myocilin |
| chr4_+_130640436 | 0.03 |
ENSMUST00000151698.8
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr7_+_45063079 | 0.03 |
ENSMUST00000058879.8
|
Ntf5
|
neurotrophin 5 |
| chr5_+_20112771 | 0.03 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_-_137309415 | 0.03 |
ENSMUST00000238941.2
|
Ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr2_+_36827133 | 0.03 |
ENSMUST00000095021.2
|
Olfr356
|
olfactory receptor 356 |
| chr1_-_180524587 | 0.03 |
ENSMUST00000027778.8
|
Mixl1
|
Mix1 homeobox-like 1 (Xenopus laevis) |
| chr14_+_53913598 | 0.03 |
ENSMUST00000103662.6
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
| chr7_+_45055077 | 0.03 |
ENSMUST00000107774.3
|
Kcna7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr5_-_49682106 | 0.02 |
ENSMUST00000176191.8
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr15_-_103275190 | 0.02 |
ENSMUST00000023128.8
|
Itga5
|
integrin alpha 5 (fibronectin receptor alpha) |
| chr3_+_63203516 | 0.02 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr1_-_87029312 | 0.02 |
ENSMUST00000113270.3
|
Alpi
|
alkaline phosphatase, intestinal |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.4 | 1.3 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.4 | 1.2 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) cell proliferation involved in heart valve development(GO:2000793) |
| 0.3 | 0.3 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 | 2.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 0.8 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 0.9 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 0.6 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.1 | 0.9 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.6 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.1 | 0.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.8 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 0.2 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 1.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 3.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.4 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 1.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 1.9 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 0.2 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.0 | 0.1 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.1 | GO:1904631 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.0 | 1.4 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 1.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.5 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 1.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 3.5 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.6 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 3.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.0 | 1.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 1.3 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 1.2 | GO:0036122 | BMP binding(GO:0036122) BMP receptor activity(GO:0098821) |
| 0.1 | 3.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.7 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.7 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.9 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 1.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.8 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.8 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 2.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |