Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
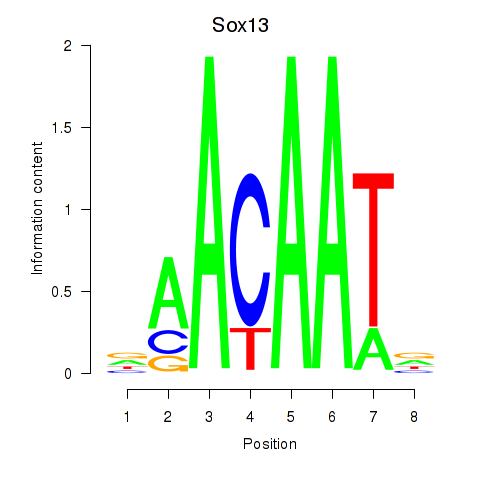
Results for Sox13
Z-value: 0.15
Transcription factors associated with Sox13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox13
|
ENSMUSG00000070643.12 | SRY (sex determining region Y)-box 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox13 | mm39_v1_chr1_-_133352115_133352138 | 0.97 | 5.3e-03 | Click! |
Activity profile of Sox13 motif
Sorted Z-values of Sox13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_44282113 | 0.11 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr8_+_108271643 | 0.05 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_172327812 | 0.05 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr11_+_29668563 | 0.04 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr18_-_43610829 | 0.04 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr2_+_4886298 | 0.03 |
ENSMUST00000027973.14
|
Sephs1
|
selenophosphate synthetase 1 |
| chr7_-_84328553 | 0.03 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr10_+_57362512 | 0.02 |
ENSMUST00000220042.2
|
Hsf2
|
heat shock factor 2 |
| chr10_+_57362457 | 0.02 |
ENSMUST00000079833.6
|
Hsf2
|
heat shock factor 2 |
| chr5_-_24782465 | 0.02 |
ENSMUST00000030795.10
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr5_-_121511476 | 0.02 |
ENSMUST00000202064.2
|
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr15_+_6416079 | 0.02 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr14_-_47805861 | 0.01 |
ENSMUST00000228784.2
ENSMUST00000042988.7 |
Atg14
|
autophagy related 14 |
| chr14_-_121935829 | 0.01 |
ENSMUST00000040700.9
ENSMUST00000212181.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr13_-_63712349 | 0.01 |
ENSMUST00000192155.6
|
Ptch1
|
patched 1 |
| chr9_-_57673128 | 0.01 |
ENSMUST00000065330.8
|
Clk3
|
CDC-like kinase 3 |
| chr4_-_58499398 | 0.01 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr8_+_3543131 | 0.01 |
ENSMUST00000061508.8
ENSMUST00000207318.2 |
Zfp358
|
zinc finger protein 358 |
| chr2_-_85992176 | 0.01 |
ENSMUST00000099908.6
ENSMUST00000215624.2 |
Olfr1042
|
olfactory receptor 1042 |
| chr14_+_5894220 | 0.01 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr8_-_62576140 | 0.01 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr16_-_59459745 | 0.01 |
ENSMUST00000099646.10
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr12_-_104439589 | 0.01 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chrX_+_139243012 | 0.01 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr11_+_73819052 | 0.00 |
ENSMUST00000205791.2
|
Olfr396-ps1
|
olfactory receptor 396, pseudogene 1 |
| chr3_-_141687987 | 0.00 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr2_-_89448589 | 0.00 |
ENSMUST00000216129.2
|
Olfr1248
|
olfactory receptor 1248 |
| chr2_+_69869459 | 0.00 |
ENSMUST00000060208.11
|
Myo3b
|
myosin IIIB |
| chr11_-_99556781 | 0.00 |
ENSMUST00000100476.3
|
Krtap4-6
|
keratin associated protein 4-6 |
| chr5_+_88523967 | 0.00 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr9_-_75504926 | 0.00 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chr8_+_106154266 | 0.00 |
ENSMUST00000067305.8
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr2_+_87405430 | 0.00 |
ENSMUST00000081034.2
|
Olfr1129
|
olfactory receptor 1129 |
| chr6_+_43150886 | 0.00 |
ENSMUST00000059512.5
|
Olfr13
|
olfactory receptor 13 |
| chr11_-_99328969 | 0.00 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr2_-_89313477 | 0.00 |
ENSMUST00000099778.2
|
Olfr1241
|
olfactory receptor 1241 |
| chr13_-_22286934 | 0.00 |
ENSMUST00000186062.3
|
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr2_-_85439252 | 0.00 |
ENSMUST00000099921.2
|
Olfr1000
|
olfactory receptor 1000 |
| chr11_-_74098115 | 0.00 |
ENSMUST00000078936.5
|
Olfr43
|
olfactory receptor 43 |
| chr18_-_42132106 | 0.00 |
ENSMUST00000097591.5
|
Grxcr2
|
glutaredoxin, cysteine rich 2 |
| chr9_-_29323500 | 0.00 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr2_-_89237422 | 0.00 |
ENSMUST00000099781.5
|
Olfr1238
|
olfactory receptor 1238 |
| chr13_-_21327251 | 0.00 |
ENSMUST00000055298.6
|
Olfr1368
|
olfactory receptor 1368 |
| chr6_+_48951872 | 0.00 |
ENSMUST00000184917.6
|
Doxl2
|
diamine oxidase-like protein 2 |
| chr6_-_69377328 | 0.00 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr6_-_93769426 | 0.00 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr6_+_131463363 | 0.00 |
ENSMUST00000159229.3
ENSMUST00000075020.10 |
Gm6619
5430401F13Rik
|
predicted gene 6619 RIKEN cDNA 5430401F13 gene |
| chr11_-_99695272 | 0.00 |
ENSMUST00000105056.2
|
Gm11554
|
predicted gene 11554 |
| chr1_+_19173246 | 0.00 |
ENSMUST00000037294.8
|
Tfap2d
|
transcription factor AP-2, delta |
| chr11_-_99706484 | 0.00 |
ENSMUST00000105055.2
|
Gm11564
|
predicted gene 11564 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |