Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
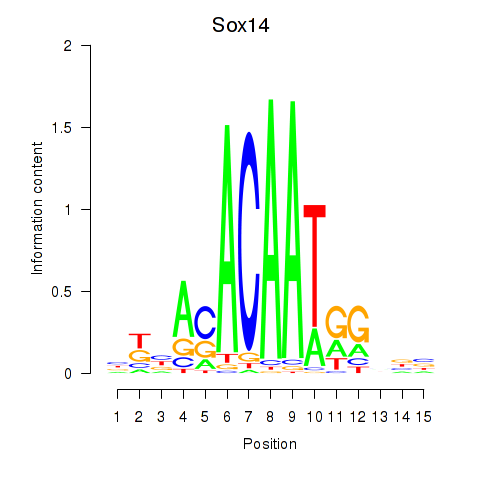
Results for Sox14
Z-value: 0.51
Transcription factors associated with Sox14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox14
|
ENSMUSG00000053747.10 | SRY (sex determining region Y)-box 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox14 | mm39_v1_chr9_-_99758200_99758245 | 0.20 | 7.4e-01 | Click! |
Activity profile of Sox14 motif
Sorted Z-values of Sox14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_23929490 | 0.63 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr5_-_110534828 | 0.40 |
ENSMUST00000198834.2
ENSMUST00000056124.11 |
Fbrsl1
|
fibrosin-like 1 |
| chr1_-_72914036 | 0.39 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr5_-_136275407 | 0.37 |
ENSMUST00000196245.2
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr13_-_22016364 | 0.32 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr13_-_21900313 | 0.31 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr16_-_58620631 | 0.29 |
ENSMUST00000206205.3
|
Olfr173
|
olfactory receptor 173 |
| chr13_+_21995906 | 0.28 |
ENSMUST00000104941.4
|
H4c17
|
H4 clustered histone 17 |
| chr1_-_85888729 | 0.26 |
ENSMUST00000086975.7
|
Gpr55
|
G protein-coupled receptor 55 |
| chr19_-_10847121 | 0.25 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr16_+_51851948 | 0.24 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr4_-_155947819 | 0.24 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr11_-_110142565 | 0.24 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr13_-_58261406 | 0.24 |
ENSMUST00000160860.9
|
Klhl3
|
kelch-like 3 |
| chr14_-_33700719 | 0.24 |
ENSMUST00000166737.2
|
Zfp488
|
zinc finger protein 488 |
| chr16_+_51851588 | 0.22 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr12_-_99359265 | 0.21 |
ENSMUST00000177451.8
|
Foxn3
|
forkhead box N3 |
| chr4_-_132149704 | 0.21 |
ENSMUST00000152271.8
ENSMUST00000084170.12 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr16_+_51851917 | 0.21 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr7_+_140261864 | 0.20 |
ENSMUST00000214637.3
|
Olfr45
|
olfactory receptor 45 |
| chr11_-_97913420 | 0.19 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr16_+_65317389 | 0.19 |
ENSMUST00000176330.8
ENSMUST00000004964.15 ENSMUST00000176038.8 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr17_-_29226886 | 0.19 |
ENSMUST00000232723.2
|
Stk38
|
serine/threonine kinase 38 |
| chr14_-_18585601 | 0.18 |
ENSMUST00000181779.2
|
Gm2916
|
predicted gene 2916 |
| chr5_-_132570710 | 0.18 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr16_-_90081300 | 0.17 |
ENSMUST00000039280.9
ENSMUST00000232371.2 |
Scaf4
|
SR-related CTD-associated factor 4 |
| chr6_+_65648574 | 0.17 |
ENSMUST00000054351.6
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr3_+_90201388 | 0.17 |
ENSMUST00000199607.5
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr17_-_34340918 | 0.17 |
ENSMUST00000151986.2
|
Brd2
|
bromodomain containing 2 |
| chr1_+_39940189 | 0.16 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr8_-_106016097 | 0.16 |
ENSMUST00000171788.8
|
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr12_-_115832846 | 0.16 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr6_-_137626207 | 0.16 |
ENSMUST00000134630.6
ENSMUST00000058210.13 ENSMUST00000111878.8 |
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr4_-_129334593 | 0.16 |
ENSMUST00000053042.6
ENSMUST00000106046.8 |
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr7_-_126992776 | 0.16 |
ENSMUST00000165495.2
ENSMUST00000106303.3 ENSMUST00000074249.7 |
E430018J23Rik
|
RIKEN cDNA E430018J23 gene |
| chr16_+_37909363 | 0.16 |
ENSMUST00000023507.13
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr9_+_72832904 | 0.16 |
ENSMUST00000038489.6
|
Pygo1
|
pygopus 1 |
| chr5_+_108213562 | 0.15 |
ENSMUST00000172045.8
|
Mtf2
|
metal response element binding transcription factor 2 |
| chr7_-_100164007 | 0.15 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr17_-_59320257 | 0.14 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr17_+_48623157 | 0.14 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr4_-_132149780 | 0.14 |
ENSMUST00000102568.10
|
Phactr4
|
phosphatase and actin regulator 4 |
| chr7_+_79675727 | 0.13 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr4_+_137944546 | 0.13 |
ENSMUST00000130071.2
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr11_+_67477347 | 0.13 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr13_-_34314328 | 0.13 |
ENSMUST00000075774.5
|
Tubb2b
|
tubulin, beta 2B class IIB |
| chr7_+_128290204 | 0.12 |
ENSMUST00000118605.2
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr6_-_97594498 | 0.12 |
ENSMUST00000113355.9
|
Frmd4b
|
FERM domain containing 4B |
| chr2_-_60711706 | 0.12 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr4_+_155685830 | 0.12 |
ENSMUST00000105608.9
|
Slc35e2
|
solute carrier family 35, member E2 |
| chr6_+_120341055 | 0.12 |
ENSMUST00000005108.10
|
Kdm5a
|
lysine (K)-specific demethylase 5A |
| chr9_-_70410611 | 0.12 |
ENSMUST00000215848.2
ENSMUST00000113595.2 ENSMUST00000213647.2 |
Rnf111
|
ring finger 111 |
| chr2_-_38955518 | 0.12 |
ENSMUST00000039165.15
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr7_+_102790086 | 0.12 |
ENSMUST00000118682.3
|
Olfr588-ps1
|
olfactory receptor 588, pseudogene 1 |
| chr18_+_37580692 | 0.12 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr3_-_57599956 | 0.12 |
ENSMUST00000238789.2
ENSMUST00000197088.5 ENSMUST00000099091.4 |
Ankub1
|
ankrin repeat and ubiquitin domain containing 1 |
| chr2_-_24809583 | 0.11 |
ENSMUST00000046227.12
ENSMUST00000114432.9 ENSMUST00000091348.11 ENSMUST00000102938.10 ENSMUST00000150379.2 ENSMUST00000152161.8 ENSMUST00000147147.8 |
Ehmt1
|
euchromatic histone methyltransferase 1 |
| chr7_+_99184858 | 0.11 |
ENSMUST00000032995.15
ENSMUST00000162404.8 |
Arrb1
|
arrestin, beta 1 |
| chr1_+_177270101 | 0.11 |
ENSMUST00000194319.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr5_+_53748323 | 0.11 |
ENSMUST00000201883.4
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_+_15785603 | 0.11 |
ENSMUST00000181786.2
|
Gm10251
|
predicted gene 10251 |
| chr9_+_61280501 | 0.11 |
ENSMUST00000162583.8
ENSMUST00000161993.8 ENSMUST00000160882.8 ENSMUST00000160724.8 ENSMUST00000162973.8 ENSMUST00000159050.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr6_-_120341304 | 0.11 |
ENSMUST00000146667.2
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr13_+_41040657 | 0.11 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr8_-_106015682 | 0.11 |
ENSMUST00000212922.2
ENSMUST00000212219.2 |
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr2_+_90770742 | 0.10 |
ENSMUST00000005643.14
ENSMUST00000111451.10 ENSMUST00000177642.8 ENSMUST00000068726.13 ENSMUST00000068747.14 |
Celf1
|
CUGBP, Elav-like family member 1 |
| chr1_+_178146689 | 0.10 |
ENSMUST00000027781.7
|
Cox20
|
cytochrome c oxidase assembly protein 20 |
| chr4_+_122910382 | 0.10 |
ENSMUST00000102649.4
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr6_+_83688213 | 0.10 |
ENSMUST00000037807.3
|
Vax2
|
ventral anterior homeobox 2 |
| chr6_-_42669963 | 0.10 |
ENSMUST00000045140.5
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr15_+_75088220 | 0.10 |
ENSMUST00000185307.7
ENSMUST00000229521.2 |
Ly6g2
|
lymphocyte antigen 6 complex, locus G2 |
| chr19_+_6413840 | 0.10 |
ENSMUST00000113488.8
ENSMUST00000113487.8 |
Sf1
|
splicing factor 1 |
| chr17_+_21031817 | 0.09 |
ENSMUST00000232810.2
ENSMUST00000233712.2 ENSMUST00000232852.2 |
Vmn1r229
|
vomeronasal 1 receptor 229 |
| chr7_-_28824440 | 0.09 |
ENSMUST00000214374.2
ENSMUST00000179893.9 ENSMUST00000032813.10 |
Ryr1
|
ryanodine receptor 1, skeletal muscle |
| chrX_+_109857866 | 0.09 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr7_+_24230063 | 0.09 |
ENSMUST00000049020.9
|
Irgq
|
immunity-related GTPase family, Q |
| chr9_-_70411000 | 0.09 |
ENSMUST00000034739.12
|
Rnf111
|
ring finger 111 |
| chr1_+_118317153 | 0.09 |
ENSMUST00000189738.7
ENSMUST00000187713.7 ENSMUST00000165223.8 ENSMUST00000189570.7 ENSMUST00000191445.7 ENSMUST00000189262.7 |
Clasp1
|
CLIP associating protein 1 |
| chr10_-_80895949 | 0.09 |
ENSMUST00000219401.2
ENSMUST00000220317.2 ENSMUST00000005067.6 ENSMUST00000218208.2 |
Sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr7_-_44711771 | 0.09 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr4_-_142939260 | 0.09 |
ENSMUST00000105778.8
ENSMUST00000134791.2 |
Prdm2
|
PR domain containing 2, with ZNF domain |
| chr1_-_64161415 | 0.09 |
ENSMUST00000135075.2
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_-_116084810 | 0.09 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr2_+_145627900 | 0.09 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr15_-_103242697 | 0.09 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr10_-_128918779 | 0.09 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr7_-_103674780 | 0.08 |
ENSMUST00000218535.2
|
Olfr640
|
olfactory receptor 640 |
| chr13_-_103233323 | 0.08 |
ENSMUST00000166336.9
ENSMUST00000239261.2 ENSMUST00000194446.7 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_78488795 | 0.08 |
ENSMUST00000170511.3
|
BC035947
|
cDNA sequence BC035947 |
| chr9_+_19716202 | 0.08 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr9_+_61280764 | 0.08 |
ENSMUST00000160541.8
ENSMUST00000161207.8 ENSMUST00000159630.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr2_+_179666744 | 0.08 |
ENSMUST00000055485.12
|
Lsm14b
|
LSM family member 14B |
| chr15_+_81469538 | 0.08 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr2_-_38955452 | 0.08 |
ENSMUST00000112850.9
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr9_+_61280890 | 0.08 |
ENSMUST00000161689.8
|
Tle3
|
transducin-like enhancer of split 3 |
| chr11_-_5049223 | 0.08 |
ENSMUST00000079949.13
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr3_+_122523219 | 0.08 |
ENSMUST00000200389.2
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr18_-_46345661 | 0.08 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr7_+_6441687 | 0.08 |
ENSMUST00000218906.2
|
Olfr1344
|
olfactory receptor 1344 |
| chr12_-_24147247 | 0.08 |
ENSMUST00000101538.5
|
9030624G23Rik
|
RIKEN cDNA 9030624G23 gene |
| chr1_-_92107971 | 0.07 |
ENSMUST00000186002.3
ENSMUST00000097644.9 |
Hdac4
|
histone deacetylase 4 |
| chr11_-_106469938 | 0.07 |
ENSMUST00000103070.3
|
Tex2
|
testis expressed gene 2 |
| chr11_-_90528888 | 0.07 |
ENSMUST00000020858.14
ENSMUST00000107875.8 ENSMUST00000107872.8 ENSMUST00000143203.8 |
Stxbp4
|
syntaxin binding protein 4 |
| chr7_+_12946947 | 0.07 |
ENSMUST00000226717.2
ENSMUST00000227176.2 ENSMUST00000227239.2 ENSMUST00000227276.2 ENSMUST00000227319.2 ENSMUST00000227390.2 ENSMUST00000228587.2 |
Vmn1r89
|
vomeronasal 1 receptor 89 |
| chr11_+_67477501 | 0.07 |
ENSMUST00000108680.2
|
Gas7
|
growth arrest specific 7 |
| chr17_-_56783462 | 0.07 |
ENSMUST00000067538.6
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr16_-_16962279 | 0.07 |
ENSMUST00000232033.2
ENSMUST00000231597.2 ENSMUST00000232540.2 |
Ccdc116
|
coiled-coil domain containing 116 |
| chr7_-_10011933 | 0.07 |
ENSMUST00000227719.2
ENSMUST00000228622.2 ENSMUST00000228086.2 |
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr7_+_64151435 | 0.07 |
ENSMUST00000032732.15
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr1_-_128520002 | 0.07 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr1_+_192855776 | 0.06 |
ENSMUST00000161235.3
ENSMUST00000160077.2 ENSMUST00000178744.2 ENSMUST00000192189.2 ENSMUST00000110831.4 ENSMUST00000191613.2 |
A130010J15Rik
|
RIKEN cDNA A130010J15 gene |
| chr5_+_150183201 | 0.06 |
ENSMUST00000087204.9
|
Fry
|
FRY microtubule binding protein |
| chr1_-_77491683 | 0.06 |
ENSMUST00000186930.2
ENSMUST00000027451.13 ENSMUST00000188797.7 |
Epha4
|
Eph receptor A4 |
| chr3_-_88366159 | 0.06 |
ENSMUST00000147200.8
ENSMUST00000169222.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr5_-_109207435 | 0.06 |
ENSMUST00000164875.3
|
Vmn2r11
|
vomeronasal 2, receptor 11 |
| chr8_+_3543131 | 0.06 |
ENSMUST00000061508.8
ENSMUST00000207318.2 |
Zfp358
|
zinc finger protein 358 |
| chr19_+_6413703 | 0.06 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr19_+_12610870 | 0.06 |
ENSMUST00000119960.2
|
Glyat
|
glycine-N-acyltransferase |
| chr17_+_6157154 | 0.06 |
ENSMUST00000149756.8
|
Tulp4
|
tubby like protein 4 |
| chr17_-_56783376 | 0.06 |
ENSMUST00000223859.2
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr17_-_48474356 | 0.06 |
ENSMUST00000027764.10
ENSMUST00000053612.14 |
A530064D06Rik
|
RIKEN cDNA A530064D06 gene |
| chr18_+_60936910 | 0.06 |
ENSMUST00000097563.9
ENSMUST00000050487.16 ENSMUST00000167610.2 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr9_-_32452885 | 0.06 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr3_-_144525255 | 0.06 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr1_-_173703424 | 0.06 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr1_-_79739469 | 0.06 |
ENSMUST00000113512.8
ENSMUST00000113513.8 ENSMUST00000113515.8 ENSMUST00000113514.8 ENSMUST00000113510.8 ENSMUST00000113511.8 ENSMUST00000048820.14 |
Wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr7_+_127344942 | 0.06 |
ENSMUST00000189562.7
ENSMUST00000186116.7 |
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr16_+_4867876 | 0.05 |
ENSMUST00000230703.2
ENSMUST00000052449.6 |
Ubn1
|
ubinuclein 1 |
| chr2_-_57014015 | 0.05 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr18_+_37575553 | 0.05 |
ENSMUST00000056915.3
|
Pcdhb13
|
protocadherin beta 13 |
| chr12_-_105651437 | 0.05 |
ENSMUST00000041055.9
|
Atg2b
|
autophagy related 2B |
| chr8_+_120301974 | 0.05 |
ENSMUST00000093100.3
|
Dnaaf1
|
dynein, axonemal assembly factor 1 |
| chr2_-_7086018 | 0.05 |
ENSMUST00000114923.3
ENSMUST00000182706.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr15_-_74753548 | 0.05 |
ENSMUST00000077004.6
|
Ly6m
|
lymphocyte antigen 6 complex, locus M |
| chr11_+_58531220 | 0.05 |
ENSMUST00000075084.5
|
Trim58
|
tripartite motif-containing 58 |
| chr18_+_31892933 | 0.05 |
ENSMUST00000115808.4
|
Ammecr1l
|
AMME chromosomal region gene 1-like |
| chr3_-_104960264 | 0.05 |
ENSMUST00000098763.7
ENSMUST00000197437.5 |
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr2_-_39080701 | 0.05 |
ENSMUST00000142872.2
ENSMUST00000038874.12 |
Scai
|
suppressor of cancer cell invasion |
| chr19_-_56810593 | 0.05 |
ENSMUST00000118592.8
|
Ccdc186
|
coiled-coil domain containing 186 |
| chr10_-_125164826 | 0.05 |
ENSMUST00000211781.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr17_+_23161958 | 0.05 |
ENSMUST00000170322.3
ENSMUST00000234787.2 |
Vmn2r113
|
vomeronasal 2, receptor 113 |
| chr2_-_34261121 | 0.05 |
ENSMUST00000127353.3
ENSMUST00000141653.3 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr2_-_102016665 | 0.05 |
ENSMUST00000111222.2
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr13_-_89890363 | 0.05 |
ENSMUST00000109543.9
ENSMUST00000159337.8 ENSMUST00000159910.8 ENSMUST00000109544.9 |
Vcan
|
versican |
| chr17_-_34834938 | 0.04 |
ENSMUST00000097345.10
ENSMUST00000015611.14 |
Egfl8
|
EGF-like domain 8 |
| chr1_-_65142521 | 0.04 |
ENSMUST00000061497.9
|
Cryga
|
crystallin, gamma A |
| chr12_-_115825934 | 0.04 |
ENSMUST00000198777.2
|
Ighv1-77
|
immunoglobulin heavy variable 1-77 |
| chr3_-_88366351 | 0.04 |
ENSMUST00000165898.8
ENSMUST00000127436.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chrX_+_135894030 | 0.04 |
ENSMUST00000033804.5
|
Zcchc18
|
zinc finger, CCHC domain containing 18 |
| chr13_-_21744063 | 0.04 |
ENSMUST00000217519.2
|
Olfr1535
|
olfactory receptor 1535 |
| chr1_+_65225834 | 0.04 |
ENSMUST00000081154.14
|
Pikfyve
|
phosphoinositide kinase, FYVE type zinc finger containing |
| chr5_+_145217272 | 0.04 |
ENSMUST00000200246.2
|
Zscan25
|
zinc finger and SCAN domain containing 25 |
| chr9_+_22010512 | 0.04 |
ENSMUST00000214601.2
ENSMUST00000001384.6 |
Cnn1
|
calponin 1 |
| chr17_-_29226965 | 0.04 |
ENSMUST00000009138.13
ENSMUST00000119274.3 |
Stk38
|
serine/threonine kinase 38 |
| chr7_+_99184645 | 0.04 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr14_-_68893253 | 0.04 |
ENSMUST00000225767.3
ENSMUST00000111072.8 ENSMUST00000022642.6 ENSMUST00000224039.2 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr3_+_65016750 | 0.04 |
ENSMUST00000049230.11
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr3_+_89622323 | 0.04 |
ENSMUST00000098924.9
|
Adar
|
adenosine deaminase, RNA-specific |
| chr3_+_60503051 | 0.04 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr10_-_63174801 | 0.04 |
ENSMUST00000020257.13
ENSMUST00000177694.8 ENSMUST00000105442.3 |
Sirt1
|
sirtuin 1 |
| chr10_-_117212826 | 0.04 |
ENSMUST00000177145.8
ENSMUST00000176670.8 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr15_-_79430742 | 0.04 |
ENSMUST00000231053.2
ENSMUST00000229431.2 |
Ddx17
|
DEAD box helicase 17 |
| chr1_+_132996237 | 0.03 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr11_-_68848271 | 0.03 |
ENSMUST00000108671.2
|
Arhgef15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr16_-_58583539 | 0.03 |
ENSMUST00000214139.2
|
Olfr172
|
olfactory receptor 172 |
| chr8_+_70275079 | 0.03 |
ENSMUST00000164890.8
ENSMUST00000034325.6 ENSMUST00000238452.2 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr2_-_102016717 | 0.03 |
ENSMUST00000058790.12
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr12_-_36092475 | 0.03 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr8_-_71848429 | 0.03 |
ENSMUST00000049184.9
|
Ushbp1
|
USH1 protein network component harmonin binding protein 1 |
| chr13_+_112937320 | 0.03 |
ENSMUST00000016144.12
|
Plpp1
|
phospholipid phosphatase 1 |
| chr11_-_78056347 | 0.03 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr2_-_154411640 | 0.03 |
ENSMUST00000000894.6
|
E2f1
|
E2F transcription factor 1 |
| chr14_-_7766684 | 0.03 |
ENSMUST00000225630.3
ENSMUST00000225979.3 ENSMUST00000226079.3 ENSMUST00000223607.3 ENSMUST00000223761.3 ENSMUST00000223981.3 ENSMUST00000224222.3 ENSMUST00000225175.3 ENSMUST00000225238.3 ENSMUST00000225232.3 |
Slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr2_-_88590348 | 0.03 |
ENSMUST00000131038.4
ENSMUST00000213138.3 |
Olfr1199
|
olfactory receptor 1199 |
| chr6_-_50238723 | 0.03 |
ENSMUST00000101405.10
ENSMUST00000165099.8 ENSMUST00000170142.8 |
Gsdme
|
gasdermin E |
| chr12_-_32258604 | 0.03 |
ENSMUST00000053215.14
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr8_+_11778039 | 0.03 |
ENSMUST00000110909.9
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr1_-_189075515 | 0.03 |
ENSMUST00000193319.6
|
Kcnk2
|
potassium channel, subfamily K, member 2 |
| chr10_-_18890281 | 0.03 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr9_+_98305014 | 0.03 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chrX_-_94240056 | 0.03 |
ENSMUST00000200628.2
ENSMUST00000197364.5 ENSMUST00000181987.8 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr11_+_9143415 | 0.03 |
ENSMUST00000151522.2
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr2_+_32766126 | 0.03 |
ENSMUST00000028135.15
|
Niban2
|
niban apoptosis regulator 2 |
| chr12_+_108572015 | 0.03 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr1_+_24717793 | 0.03 |
ENSMUST00000186190.2
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr16_+_31482658 | 0.03 |
ENSMUST00000115201.8
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr14_+_55855484 | 0.03 |
ENSMUST00000002395.8
|
Rec8
|
REC8 meiotic recombination protein |
| chr4_+_117706390 | 0.03 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr1_+_88154727 | 0.03 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr1_+_65225886 | 0.03 |
ENSMUST00000097707.5
|
Pikfyve
|
phosphoinositide kinase, FYVE type zinc finger containing |
| chr9_-_88364593 | 0.03 |
ENSMUST00000173801.8
ENSMUST00000069221.12 ENSMUST00000172508.2 |
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr19_-_4993060 | 0.02 |
ENSMUST00000133504.2
ENSMUST00000133254.2 ENSMUST00000120475.8 ENSMUST00000025834.15 |
Peli3
|
pellino 3 |
| chr12_-_99849660 | 0.02 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr7_+_121818692 | 0.02 |
ENSMUST00000033152.5
|
Chp2
|
calcineurin-like EF hand protein 2 |
| chr11_+_110888313 | 0.02 |
ENSMUST00000106635.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_-_37535170 | 0.02 |
ENSMUST00000148047.2
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr1_+_191638854 | 0.02 |
ENSMUST00000044954.7
|
Slc30a1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr9_+_123901979 | 0.02 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr14_+_76082533 | 0.02 |
ENSMUST00000110894.9
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr17_-_34834745 | 0.02 |
ENSMUST00000168353.4
|
Egfl8
|
EGF-like domain 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.7 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.2 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.4 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.1 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.0 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.0 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.0 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |