Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
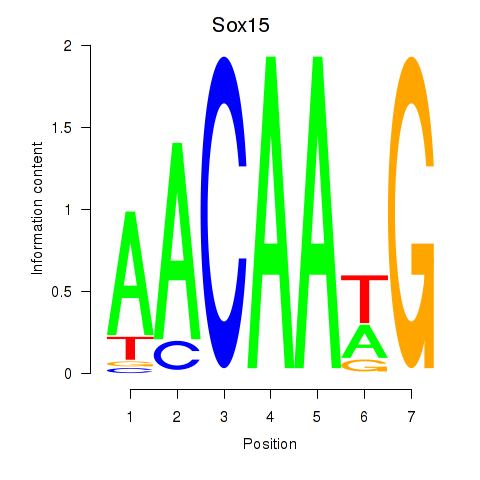
Results for Sox15
Z-value: 1.29
Transcription factors associated with Sox15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox15
|
ENSMUSG00000041287.6 | SRY (sex determining region Y)-box 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox15 | mm39_v1_chr11_+_69546140_69546140 | 0.26 | 6.7e-01 | Click! |
Activity profile of Sox15 motif
Sorted Z-values of Sox15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_121220146 | 1.45 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr6_-_52217821 | 1.08 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr12_+_24758724 | 0.96 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr12_+_24758968 | 0.93 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr10_-_37014859 | 0.86 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr16_+_16964801 | 0.82 |
ENSMUST00000232479.2
ENSMUST00000232344.2 ENSMUST00000069064.7 |
Ydjc
|
YdjC homolog (bacterial) |
| chr11_+_87651359 | 0.82 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr13_+_94954202 | 0.66 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr1_+_86454431 | 0.65 |
ENSMUST00000045897.15
ENSMUST00000186255.7 ENSMUST00000188699.7 |
Ptma
|
prothymosin alpha |
| chr15_+_79543397 | 0.64 |
ENSMUST00000023064.9
|
Cby1
|
chibby family member 1, beta catenin antagonist |
| chr13_+_9326513 | 0.58 |
ENSMUST00000174552.8
|
Dip2c
|
disco interacting protein 2 homolog C |
| chr11_-_106605772 | 0.57 |
ENSMUST00000124958.3
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr19_+_44282113 | 0.53 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr19_-_41836514 | 0.53 |
ENSMUST00000059231.4
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr9_+_13677266 | 0.52 |
ENSMUST00000152532.8
|
Mtmr2
|
myotubularin related protein 2 |
| chr1_+_86454511 | 0.49 |
ENSMUST00000188533.2
|
Ptma
|
prothymosin alpha |
| chr6_-_47790272 | 0.47 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr11_+_93935021 | 0.47 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr9_+_35179153 | 0.45 |
ENSMUST00000034543.5
|
Rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr11_+_93935066 | 0.45 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr16_+_93404719 | 0.43 |
ENSMUST00000039659.9
ENSMUST00000231762.2 |
Cbr1
|
carbonyl reductase 1 |
| chr18_+_11766333 | 0.43 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr19_+_5742880 | 0.42 |
ENSMUST00000235661.2
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr10_+_53472853 | 0.39 |
ENSMUST00000219271.2
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr6_-_86710250 | 0.37 |
ENSMUST00000001185.14
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr15_+_6416079 | 0.37 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr8_-_86281946 | 0.37 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr1_+_172327812 | 0.37 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr3_+_32763313 | 0.37 |
ENSMUST00000126144.3
|
Actl6a
|
actin-like 6A |
| chr10_+_53473032 | 0.35 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr17_-_71158052 | 0.35 |
ENSMUST00000186358.6
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr2_-_84255602 | 0.34 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr11_+_93935156 | 0.34 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr1_-_46927230 | 0.33 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr5_+_36050663 | 0.32 |
ENSMUST00000064571.11
|
Afap1
|
actin filament associated protein 1 |
| chr17_-_71158184 | 0.32 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr4_+_128582519 | 0.29 |
ENSMUST00000106080.8
|
Phc2
|
polyhomeotic 2 |
| chr19_-_7218363 | 0.28 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr13_-_43632368 | 0.28 |
ENSMUST00000222651.2
|
Ranbp9
|
RAN binding protein 9 |
| chr4_+_102427247 | 0.27 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr8_+_22996233 | 0.27 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr15_-_37008011 | 0.26 |
ENSMUST00000226671.2
|
Zfp706
|
zinc finger protein 706 |
| chr2_+_160573604 | 0.26 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr5_+_148202011 | 0.25 |
ENSMUST00000110515.9
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr10_+_79986280 | 0.24 |
ENSMUST00000153477.8
|
Midn
|
midnolin |
| chr11_+_61544085 | 0.23 |
ENSMUST00000004959.3
|
Grap
|
GRB2-related adaptor protein |
| chr12_-_83643883 | 0.23 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr5_-_24782465 | 0.23 |
ENSMUST00000030795.10
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr7_+_65759198 | 0.23 |
ENSMUST00000036372.8
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr16_-_45940602 | 0.22 |
ENSMUST00000023336.10
|
Cd96
|
CD96 antigen |
| chr5_+_148202117 | 0.22 |
ENSMUST00000110514.8
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr14_-_71003973 | 0.22 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr3_+_85946145 | 0.21 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr9_-_35179042 | 0.21 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chr10_+_87926932 | 0.20 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr5_+_148202075 | 0.20 |
ENSMUST00000071878.12
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr1_+_87332638 | 0.20 |
ENSMUST00000173152.2
ENSMUST00000173663.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr9_+_54771064 | 0.19 |
ENSMUST00000034843.9
|
Ireb2
|
iron responsive element binding protein 2 |
| chr19_-_7218512 | 0.19 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr9_+_96141299 | 0.18 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chrX_-_56438322 | 0.18 |
ENSMUST00000114730.8
|
Rbmx
|
RNA binding motif protein, X chromosome |
| chr3_-_9029097 | 0.18 |
ENSMUST00000091354.12
ENSMUST00000094381.11 |
Tpd52
|
tumor protein D52 |
| chr9_+_96141317 | 0.18 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr10_+_17672004 | 0.18 |
ENSMUST00000037964.7
|
Txlnb
|
taxilin beta |
| chr2_-_164876690 | 0.17 |
ENSMUST00000122070.2
ENSMUST00000121377.8 ENSMUST00000153905.2 ENSMUST00000040381.15 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr11_-_106606076 | 0.17 |
ENSMUST00000080853.11
ENSMUST00000183610.8 ENSMUST00000103069.10 ENSMUST00000106796.9 |
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chrX_-_56438380 | 0.17 |
ENSMUST00000143310.2
ENSMUST00000098470.9 ENSMUST00000114726.8 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr13_-_48746836 | 0.17 |
ENSMUST00000238995.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr18_+_34758062 | 0.16 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A |
| chr13_-_59930059 | 0.16 |
ENSMUST00000225581.2
|
Gm49354
|
predicted gene, 49354 |
| chr6_-_148846247 | 0.16 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr9_-_64928927 | 0.16 |
ENSMUST00000036615.7
|
Hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr8_-_87472562 | 0.16 |
ENSMUST00000045296.6
|
Siah1a
|
siah E3 ubiquitin protein ligase 1A |
| chr11_+_93934940 | 0.15 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr9_+_96140781 | 0.15 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr7_-_19595221 | 0.15 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr6_+_86605146 | 0.14 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr9_-_106666329 | 0.14 |
ENSMUST00000046502.7
|
Rad54l2
|
RAD54 like 2 (S. cerevisiae) |
| chr11_+_96214078 | 0.14 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr18_-_34757653 | 0.13 |
ENSMUST00000003876.10
ENSMUST00000115766.8 ENSMUST00000097626.10 ENSMUST00000115765.2 |
Brd8
|
bromodomain containing 8 |
| chr18_+_34757687 | 0.13 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A |
| chr11_-_78388560 | 0.13 |
ENSMUST00000061174.7
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr6_+_65019574 | 0.12 |
ENSMUST00000031984.9
ENSMUST00000205118.3 |
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr12_-_83643964 | 0.12 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chrX_+_70600481 | 0.11 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chr11_-_78388284 | 0.11 |
ENSMUST00000108287.10
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr5_+_92285748 | 0.11 |
ENSMUST00000031355.10
ENSMUST00000202155.2 |
Uso1
|
USO1 vesicle docking factor |
| chr8_+_11362805 | 0.10 |
ENSMUST00000033899.14
|
Col4a2
|
collagen, type IV, alpha 2 |
| chr5_+_20112500 | 0.10 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_+_180158035 | 0.09 |
ENSMUST00000070181.7
|
Itpkb
|
inositol 1,4,5-trisphosphate 3-kinase B |
| chr6_+_86381201 | 0.09 |
ENSMUST00000095754.10
ENSMUST00000095753.9 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr6_-_52141796 | 0.08 |
ENSMUST00000014848.11
|
Hoxa2
|
homeobox A2 |
| chr14_+_79689230 | 0.08 |
ENSMUST00000100359.3
ENSMUST00000226192.2 |
Kbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr10_-_89270554 | 0.08 |
ENSMUST00000220071.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr2_-_6726701 | 0.07 |
ENSMUST00000114927.9
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr18_+_34757666 | 0.07 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr2_-_6726998 | 0.07 |
ENSMUST00000182657.2
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr1_-_16589511 | 0.07 |
ENSMUST00000162751.8
ENSMUST00000027052.13 ENSMUST00000149320.9 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr18_+_37813286 | 0.06 |
ENSMUST00000192931.2
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1 |
| chr11_+_97554192 | 0.06 |
ENSMUST00000044730.12
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 6 |
| chr14_+_5894220 | 0.06 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr5_+_67125759 | 0.06 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr10_+_57362512 | 0.06 |
ENSMUST00000220042.2
|
Hsf2
|
heat shock factor 2 |
| chr14_-_52151537 | 0.06 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr9_-_103357564 | 0.06 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr4_+_5724305 | 0.05 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr10_-_89270527 | 0.05 |
ENSMUST00000218764.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr2_-_170248421 | 0.05 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr12_-_119202527 | 0.04 |
ENSMUST00000026360.9
|
Itgb8
|
integrin beta 8 |
| chr11_+_118913788 | 0.04 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr18_+_37130860 | 0.04 |
ENSMUST00000115659.6
|
Pcdha9
|
protocadherin alpha 9 |
| chr4_+_19280850 | 0.04 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr7_+_73025243 | 0.04 |
ENSMUST00000119206.3
ENSMUST00000094312.12 |
Rgma
|
repulsive guidance molecule family member A |
| chr6_-_128558560 | 0.04 |
ENSMUST00000060574.9
|
A2ml1
|
alpha-2-macroglobulin like 1 |
| chr17_-_78991691 | 0.04 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr2_-_65397809 | 0.04 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr14_-_80008745 | 0.03 |
ENSMUST00000039568.11
ENSMUST00000195355.2 |
Pcdh8
|
protocadherin 8 |
| chr6_-_138399896 | 0.03 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr7_+_126359763 | 0.03 |
ENSMUST00000091328.4
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr5_+_88712840 | 0.03 |
ENSMUST00000196894.5
ENSMUST00000198965.5 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr8_-_122379631 | 0.03 |
ENSMUST00000046386.5
|
Zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr6_-_86710220 | 0.03 |
ENSMUST00000113679.2
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr5_+_20112704 | 0.03 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr8_+_45960931 | 0.03 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_-_113911640 | 0.03 |
ENSMUST00000101044.9
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr11_-_99884818 | 0.02 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr10_+_52267702 | 0.02 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr16_-_28748410 | 0.02 |
ENSMUST00000100023.3
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr18_+_37440497 | 0.02 |
ENSMUST00000056712.4
|
Pcdhb4
|
protocadherin beta 4 |
| chr3_-_33136153 | 0.02 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr14_-_48900192 | 0.02 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr11_-_99742434 | 0.02 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr19_+_22670134 | 0.02 |
ENSMUST00000237470.2
ENSMUST00000099564.10 ENSMUST00000099569.10 ENSMUST00000099566.5 ENSMUST00000235712.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr6_+_86381236 | 0.02 |
ENSMUST00000095752.9
ENSMUST00000130967.8 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr1_-_134163102 | 0.02 |
ENSMUST00000187631.2
ENSMUST00000038191.8 ENSMUST00000086465.6 |
Adora1
|
adenosine A1 receptor |
| chr2_+_89832691 | 0.02 |
ENSMUST00000061701.5
|
Olfr1262
|
olfactory receptor 1262 |
| chr9_-_75504926 | 0.02 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chr14_-_109151590 | 0.02 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr10_+_87695117 | 0.02 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr1_-_138770167 | 0.02 |
ENSMUST00000112026.4
ENSMUST00000019374.14 |
Lhx9
|
LIM homeobox protein 9 |
| chr10_+_129406526 | 0.02 |
ENSMUST00000059957.4
|
Olfr794
|
olfactory receptor 794 |
| chr5_-_134975773 | 0.02 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr1_+_158190090 | 0.02 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chrX_-_58612709 | 0.02 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr16_+_25620652 | 0.02 |
ENSMUST00000115304.8
ENSMUST00000115305.2 ENSMUST00000040231.13 ENSMUST00000115306.8 |
Trp63
|
transformation related protein 63 |
| chr10_+_57362457 | 0.02 |
ENSMUST00000079833.6
|
Hsf2
|
heat shock factor 2 |
| chr15_-_65784103 | 0.01 |
ENSMUST00000079776.14
|
Oc90
|
otoconin 90 |
| chr3_-_127631068 | 0.01 |
ENSMUST00000051737.8
ENSMUST00000200409.5 |
Ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_-_89774457 | 0.01 |
ENSMUST00000090695.3
|
Olfr1259
|
olfactory receptor 1259 |
| chr10_+_29019645 | 0.01 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr5_-_134944366 | 0.01 |
ENSMUST00000008987.5
|
Cldn13
|
claudin 13 |
| chr3_+_31956656 | 0.01 |
ENSMUST00000119310.8
|
Kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr15_-_65784246 | 0.01 |
ENSMUST00000060522.11
|
Oc90
|
otoconin 90 |
| chr2_+_3115250 | 0.01 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr5_+_67125902 | 0.01 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr1_+_31215482 | 0.01 |
ENSMUST00000062560.14
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chrX_+_142301666 | 0.01 |
ENSMUST00000134402.8
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr1_+_158189900 | 0.01 |
ENSMUST00000170718.7
|
Astn1
|
astrotactin 1 |
| chr6_-_93889483 | 0.01 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_+_19282278 | 0.01 |
ENSMUST00000064976.6
|
Tfap2b
|
transcription factor AP-2 beta |
| chr11_+_67061837 | 0.01 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr3_+_31956814 | 0.01 |
ENSMUST00000192429.6
ENSMUST00000191869.6 ENSMUST00000178668.2 ENSMUST00000119970.8 |
Kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr1_+_158189831 | 0.01 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr11_-_3813895 | 0.01 |
ENSMUST00000070552.14
|
Osbp2
|
oxysterol binding protein 2 |
| chr18_+_65209103 | 0.01 |
ENSMUST00000236764.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr8_+_45960855 | 0.00 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr17_+_52909721 | 0.00 |
ENSMUST00000039366.11
|
Kcnh8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chrX_+_111513971 | 0.00 |
ENSMUST00000071814.13
|
Zfp711
|
zinc finger protein 711 |
| chr5_+_30868908 | 0.00 |
ENSMUST00000114729.8
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr13_-_54897425 | 0.00 |
ENSMUST00000099506.2
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr3_-_64194838 | 0.00 |
ENSMUST00000170244.3
|
Vmn2r3
|
vomeronasal 2, receptor 3 |
| chr13_-_54897660 | 0.00 |
ENSMUST00000135343.2
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chrX_+_111510223 | 0.00 |
ENSMUST00000113409.8
|
Zfp711
|
zinc finger protein 711 |
| chrX_+_139243012 | 0.00 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr10_+_87695352 | 0.00 |
ENSMUST00000121952.8
ENSMUST00000126490.8 |
Igf1
|
insulin-like growth factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.2 | 1.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 0.5 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.7 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.5 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.3 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 1.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.5 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.2 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.3 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.9 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0097346 | Swr1 complex(GO:0000812) INO80-type complex(GO:0097346) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.8 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 1.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.4 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.5 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |