Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Sox17
Z-value: 0.63
Transcription factors associated with Sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox17
|
ENSMUSG00000025902.14 | SRY (sex determining region Y)-box 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox17 | mm39_v1_chr1_-_4567577_4567614 | -0.99 | 1.7e-03 | Click! |
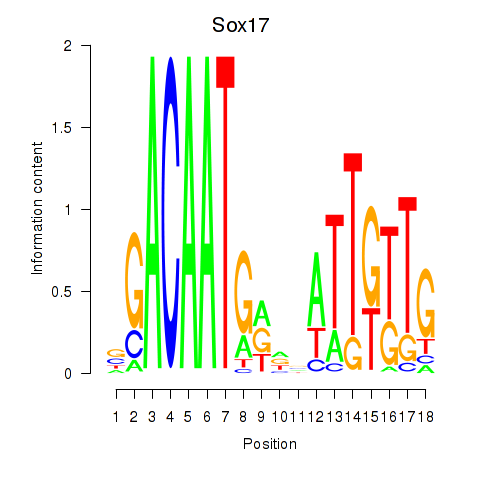
Activity profile of Sox17 motif
Sorted Z-values of Sox17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_51181956 | 0.39 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr15_+_85090150 | 0.35 |
ENSMUST00000057410.14
ENSMUST00000109432.4 |
Fbln1
|
fibulin 1 |
| chr7_+_97049210 | 0.33 |
ENSMUST00000032882.9
ENSMUST00000149122.2 |
Ndufc2
|
NADH:ubiquinone oxidoreductase subunit C2 |
| chr13_+_54225828 | 0.31 |
ENSMUST00000021930.10
|
Sfxn1
|
sideroflexin 1 |
| chr17_-_65901946 | 0.27 |
ENSMUST00000232686.2
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr1_+_52047368 | 0.25 |
ENSMUST00000027277.7
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr9_+_57428490 | 0.23 |
ENSMUST00000000090.8
|
Cox5a
|
cytochrome c oxidase subunit 5A |
| chr19_-_32717166 | 0.23 |
ENSMUST00000235142.2
ENSMUST00000070210.6 ENSMUST00000236011.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr6_-_129484070 | 0.22 |
ENSMUST00000183258.8
ENSMUST00000182784.4 ENSMUST00000032265.13 ENSMUST00000162815.2 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr19_-_32717138 | 0.21 |
ENSMUST00000236985.2
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr2_-_105229653 | 0.21 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr10_+_93289264 | 0.19 |
ENSMUST00000016033.9
|
Lta4h
|
leukotriene A4 hydrolase |
| chr15_-_74855264 | 0.19 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr17_-_23783063 | 0.18 |
ENSMUST00000095606.5
|
Zfp213
|
zinc finger protein 213 |
| chr15_+_74593041 | 0.18 |
ENSMUST00000070923.3
|
Them6
|
thioesterase superfamily member 6 |
| chr14_+_30856687 | 0.18 |
ENSMUST00000090212.5
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr10_+_77891161 | 0.16 |
ENSMUST00000131825.8
ENSMUST00000139539.8 ENSMUST00000123940.2 |
Dnmt3l
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr17_+_8529932 | 0.16 |
ENSMUST00000154553.2
ENSMUST00000140890.3 |
Sft2d1
Gm49987
|
SFT2 domain containing 1 predicted gene, 49987 |
| chr9_+_66853343 | 0.16 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr5_-_107745443 | 0.15 |
ENSMUST00000100949.10
ENSMUST00000078021.13 |
Glmn
|
glomulin, FKBP associated protein |
| chr10_+_80662490 | 0.15 |
ENSMUST00000060987.15
ENSMUST00000177850.8 ENSMUST00000180036.8 ENSMUST00000179172.8 |
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr7_+_28937859 | 0.15 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr18_-_33346885 | 0.15 |
ENSMUST00000025236.9
|
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr10_-_117215920 | 0.15 |
ENSMUST00000175924.2
|
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr16_-_18232202 | 0.13 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chr5_-_115410941 | 0.13 |
ENSMUST00000040555.15
ENSMUST00000112096.9 ENSMUST00000112097.8 |
Rnf10
|
ring finger protein 10 |
| chr14_+_73379930 | 0.13 |
ENSMUST00000170370.8
ENSMUST00000164822.8 ENSMUST00000165429.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr18_+_35731735 | 0.12 |
ENSMUST00000236868.2
|
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr11_-_59340739 | 0.12 |
ENSMUST00000136436.2
ENSMUST00000150297.2 ENSMUST00000010038.10 ENSMUST00000156146.8 ENSMUST00000132969.8 ENSMUST00000120940.8 |
Snap47
|
synaptosomal-associated protein, 47 |
| chr8_-_106022676 | 0.12 |
ENSMUST00000057855.4
|
Exoc3l
|
exocyst complex component 3-like |
| chr12_-_84447702 | 0.12 |
ENSMUST00000122194.8
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr7_+_28937746 | 0.12 |
ENSMUST00000108238.8
ENSMUST00000032809.10 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr13_-_35211060 | 0.11 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr12_-_84447625 | 0.11 |
ENSMUST00000117286.2
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr1_-_182169150 | 0.11 |
ENSMUST00000051431.10
|
Fbxo28
|
F-box protein 28 |
| chr5_+_138185747 | 0.11 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr5_-_130031681 | 0.11 |
ENSMUST00000111308.8
ENSMUST00000111307.2 |
Gusb
|
glucuronidase, beta |
| chr18_-_36330367 | 0.11 |
ENSMUST00000115713.9
|
Nrg2
|
neuregulin 2 |
| chr18_-_33346819 | 0.11 |
ENSMUST00000119991.8
ENSMUST00000118990.2 |
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr7_+_28937898 | 0.11 |
ENSMUST00000138128.3
ENSMUST00000142519.3 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr5_-_107745404 | 0.11 |
ENSMUST00000124140.2
|
Glmn
|
glomulin, FKBP associated protein |
| chr2_-_15054065 | 0.11 |
ENSMUST00000028034.15
ENSMUST00000114713.2 |
Nsun6
|
NOL1/NOP2/Sun domain family member 6 |
| chr19_+_6356486 | 0.11 |
ENSMUST00000025681.8
|
Cdc42bpg
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chr5_-_137856280 | 0.10 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr7_-_143102792 | 0.10 |
ENSMUST00000072727.7
ENSMUST00000207948.2 ENSMUST00000208190.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr5_-_104169696 | 0.10 |
ENSMUST00000119025.2
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr7_-_143102524 | 0.09 |
ENSMUST00000208093.2
ENSMUST00000209098.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr18_+_35731658 | 0.09 |
ENSMUST00000041314.17
ENSMUST00000236666.2 ENSMUST00000236020.2 ENSMUST00000235400.2 |
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr11_-_51891575 | 0.09 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr9_+_21249118 | 0.09 |
ENSMUST00000034697.8
|
Slc44a2
|
solute carrier family 44, member 2 |
| chr4_-_133005039 | 0.09 |
ENSMUST00000105907.9
|
Tmem222
|
transmembrane protein 222 |
| chr1_+_135656885 | 0.09 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr8_-_112580910 | 0.08 |
ENSMUST00000034432.7
|
Cfdp1
|
craniofacial development protein 1 |
| chr16_+_8288627 | 0.08 |
ENSMUST00000046470.16
ENSMUST00000150790.2 ENSMUST00000142899.2 |
Mettl22
|
methyltransferase like 22 |
| chr6_-_148846247 | 0.08 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr5_-_125466240 | 0.08 |
ENSMUST00000108707.3
|
Ubc
|
ubiquitin C |
| chr11_+_78237492 | 0.07 |
ENSMUST00000100755.4
|
Unc119
|
unc-119 lipid binding chaperone |
| chr10_+_80084955 | 0.07 |
ENSMUST00000105364.8
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr9_+_3335135 | 0.07 |
ENSMUST00000212294.2
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr2_-_126975804 | 0.07 |
ENSMUST00000110387.4
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr14_-_121935829 | 0.06 |
ENSMUST00000040700.9
ENSMUST00000212181.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr8_-_41507808 | 0.06 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr18_+_35732272 | 0.06 |
ENSMUST00000235210.2
|
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr10_+_81656961 | 0.06 |
ENSMUST00000099439.4
|
Gm4767
|
predicted gene 4767 |
| chr9_+_3335507 | 0.05 |
ENSMUST00000212154.2
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr1_+_171330978 | 0.05 |
ENSMUST00000081527.2
|
Alyref2
|
Aly/REF export factor 2 |
| chr1_+_117855745 | 0.05 |
ENSMUST00000191265.2
|
Gm28168
|
predicted gene 28168 |
| chr2_-_89273344 | 0.05 |
ENSMUST00000216123.2
|
Olfr1240
|
olfactory receptor 1240 |
| chr17_-_21110913 | 0.05 |
ENSMUST00000061278.2
|
Vmn1r231
|
vomeronasal 1 receptor 231 |
| chr6_-_83010402 | 0.05 |
ENSMUST00000089651.6
|
Dok1
|
docking protein 1 |
| chr5_-_38659449 | 0.05 |
ENSMUST00000005238.13
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr8_-_93924426 | 0.05 |
ENSMUST00000034172.8
|
Ces1d
|
carboxylesterase 1D |
| chr13_+_97203774 | 0.05 |
ENSMUST00000169863.9
|
Fam169a
|
family with sequence similarity 169, member A |
| chr10_-_128012336 | 0.05 |
ENSMUST00000092033.4
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr3_-_153430741 | 0.05 |
ENSMUST00000064460.7
ENSMUST00000200397.5 |
St6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr11_+_11439266 | 0.04 |
ENSMUST00000109678.3
|
Spata48
|
spermatogenesis associated 48 |
| chr8_-_83129134 | 0.04 |
ENSMUST00000209363.2
|
Il15
|
interleukin 15 |
| chr8_-_61407760 | 0.04 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr19_+_34560922 | 0.04 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr6_-_129761233 | 0.04 |
ENSMUST00000118532.8
|
Klrh1
|
killer cell lectin-like receptor subfamily H, member 1 |
| chr1_+_117708863 | 0.04 |
ENSMUST00000188801.2
|
B020011L13Rik
|
RIKEN cDNA B020011L13 gene |
| chrX_+_47783881 | 0.03 |
ENSMUST00000033433.3
|
Rbmx2
|
RNA binding motif protein, X-linked 2 |
| chr15_-_97808502 | 0.03 |
ENSMUST00000173104.8
ENSMUST00000174633.8 |
Vdr
|
vitamin D (1,25-dihydroxyvitamin D3) receptor |
| chr2_-_66240408 | 0.03 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr11_-_50718508 | 0.03 |
ENSMUST00000109135.9
|
Zfp354c
|
zinc finger protein 354C |
| chr7_-_29898236 | 0.03 |
ENSMUST00000001845.13
|
Capns1
|
calpain, small subunit 1 |
| chr11_-_106278892 | 0.03 |
ENSMUST00000106813.9
ENSMUST00000141146.2 |
Icam2
|
intercellular adhesion molecule 2 |
| chr8_-_25215778 | 0.03 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr5_-_53370761 | 0.03 |
ENSMUST00000031090.8
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr12_+_51395047 | 0.03 |
ENSMUST00000119211.8
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr2_-_111100733 | 0.03 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr11_-_11920540 | 0.03 |
ENSMUST00000109653.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr10_-_10348058 | 0.03 |
ENSMUST00000179956.8
ENSMUST00000208717.2 ENSMUST00000172530.8 ENSMUST00000132573.2 |
Adgb
|
androglobin |
| chr13_-_74642055 | 0.03 |
ENSMUST00000202645.4
ENSMUST00000221173.2 |
Zfp825
|
zinc finger protein 825 |
| chr12_-_114321838 | 0.03 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr8_-_94825556 | 0.03 |
ENSMUST00000034206.6
|
Bbs2
|
Bardet-Biedl syndrome 2 (human) |
| chr2_+_36415010 | 0.03 |
ENSMUST00000216275.2
|
Olfr342
|
olfactory receptor 342 |
| chr2_+_52747855 | 0.03 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr7_+_126575510 | 0.03 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr18_+_60693797 | 0.02 |
ENSMUST00000025506.7
|
Rbm22
|
RNA binding motif protein 22 |
| chr10_+_127561259 | 0.02 |
ENSMUST00000026466.5
|
Tac2
|
tachykinin 2 |
| chr2_+_80447389 | 0.02 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr5_+_90708962 | 0.02 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr2_-_87735503 | 0.02 |
ENSMUST00000215862.2
ENSMUST00000215017.2 |
Olfr1154
|
olfactory receptor 1154 |
| chr4_-_3185358 | 0.02 |
ENSMUST00000105159.5
|
Vmn1r3
|
vomeronasal 1 receptor 3 |
| chr6_-_57956287 | 0.02 |
ENSMUST00000228585.2
|
Vmn1r25
|
vomeronasal 1 receptor 25 |
| chr11_-_68291638 | 0.02 |
ENSMUST00000108674.9
|
Ntn1
|
netrin 1 |
| chr17_-_40553176 | 0.02 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr11_-_109986763 | 0.02 |
ENSMUST00000046223.14
ENSMUST00000106664.10 ENSMUST00000106662.2 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr17_-_37481536 | 0.02 |
ENSMUST00000174673.3
|
Olfr753-ps1
|
olfactory receptor 753, pseudogene 1 |
| chr6_-_128503666 | 0.02 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr2_-_110781268 | 0.02 |
ENSMUST00000099623.10
|
Ano3
|
anoctamin 3 |
| chr7_+_11910120 | 0.02 |
ENSMUST00000062811.6
|
Vmn1r79
|
vomeronasal 1 receptor 79 |
| chr2_-_86372824 | 0.02 |
ENSMUST00000216480.2
ENSMUST00000111582.3 |
Olfr1079
|
olfactory receptor 1079 |
| chr7_+_17799863 | 0.02 |
ENSMUST00000108487.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr18_+_37440497 | 0.02 |
ENSMUST00000056712.4
|
Pcdhb4
|
protocadherin beta 4 |
| chr7_-_107424741 | 0.02 |
ENSMUST00000213252.2
|
Olfr469
|
olfactory receptor 469 |
| chr2_+_36621914 | 0.02 |
ENSMUST00000216882.2
|
Olfr347
|
olfactory receptor 347 |
| chr16_+_22769822 | 0.02 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr17_+_33487696 | 0.02 |
ENSMUST00000067840.7
|
Olfr63
|
olfactory receptor 63 |
| chr7_-_26311589 | 0.02 |
ENSMUST00000171039.2
|
Vmn1r185
|
vomeronasal 1 receptor 185 |
| chr9_+_19227643 | 0.02 |
ENSMUST00000220268.2
|
Olfr844
|
olfactory receptor 844 |
| chr11_-_109986804 | 0.01 |
ENSMUST00000100287.9
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr6_+_121709891 | 0.01 |
ENSMUST00000204124.2
|
Gm7298
|
predicted gene 7298 |
| chr8_-_93956143 | 0.01 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr5_-_86616849 | 0.01 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr13_+_19374502 | 0.01 |
ENSMUST00000198330.2
ENSMUST00000103555.3 |
Trgv6
|
T cell receptor gamma, variable 6 |
| chr16_+_38182569 | 0.01 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr7_-_79115915 | 0.01 |
ENSMUST00000073889.14
|
Polg
|
polymerase (DNA directed), gamma |
| chr16_-_22847808 | 0.01 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr17_+_38007455 | 0.01 |
ENSMUST00000213732.2
ENSMUST00000080483.10 |
Olfr119
|
olfactory receptor 119 |
| chr9_-_75504926 | 0.01 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chr7_-_19856504 | 0.01 |
ENSMUST00000079099.2
|
Vmn1r240
|
vomeronasal 1 receptor 240 |
| chr5_+_31265651 | 0.01 |
ENSMUST00000201740.4
ENSMUST00000066544.5 |
Dnajc5g
|
DnaJ heat shock protein family (Hsp40) member C5 gamma |
| chr2_+_111581351 | 0.01 |
ENSMUST00000207590.4
|
Olfr1301
|
olfactory receptor 1301 |
| chr1_+_117624861 | 0.01 |
ENSMUST00000186341.2
|
Gm28363
|
predicted gene 28363 |
| chr12_-_8589545 | 0.01 |
ENSMUST00000095863.10
ENSMUST00000165657.3 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr7_-_19902302 | 0.01 |
ENSMUST00000164526.2
|
Vmn1r94
|
vomeronasal 1 receptor 94 |
| chr12_-_40249314 | 0.01 |
ENSMUST00000095760.3
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr17_+_38059746 | 0.01 |
ENSMUST00000074555.8
ENSMUST00000174675.4 |
Olfr121
|
olfactory receptor 121 |
| chr8_+_107877252 | 0.01 |
ENSMUST00000034400.5
|
Cyb5b
|
cytochrome b5 type B |
| chr6_-_57986187 | 0.01 |
ENSMUST00000049694.4
|
Vmn1r26
|
vomeronasal 1 receptor 26 |
| chr9_-_48516447 | 0.01 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr9_+_38019984 | 0.01 |
ENSMUST00000075228.2
|
Olfr888
|
olfactory receptor 888 |
| chr7_-_103147246 | 0.01 |
ENSMUST00000216360.2
ENSMUST00000216075.2 |
Olfr609
|
olfactory receptor 609 |
| chr6_+_89717748 | 0.01 |
ENSMUST00000204949.3
ENSMUST00000228068.2 ENSMUST00000227630.2 ENSMUST00000226337.2 ENSMUST00000227039.2 ENSMUST00000226562.2 |
Vmn1r41
|
vomeronasal 1 receptor 41 |
| chr13_+_33668761 | 0.01 |
ENSMUST00000081927.4
|
Serpinb9g
|
serine (or cysteine) peptidase inhibitor, clade B, member 9g |
| chr4_+_42612121 | 0.01 |
ENSMUST00000178168.3
|
Gm10591
|
predicted gene 10591 |
| chr4_+_42255693 | 0.01 |
ENSMUST00000178864.3
|
Ccl21b
|
chemokine (C-C motif) ligand 21B (leucine) |
| chr17_+_38079498 | 0.01 |
ENSMUST00000172582.5
|
Olfr122
|
olfactory receptor 122 |
| chr12_+_51395154 | 0.01 |
ENSMUST00000121521.2
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr16_-_22847829 | 0.01 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr11_-_74098115 | 0.01 |
ENSMUST00000078936.5
|
Olfr43
|
olfactory receptor 43 |
| chrX_+_73171069 | 0.01 |
ENSMUST00000033771.11
ENSMUST00000101457.10 |
Opn1mw
|
opsin 1 (cone pigments), medium-wave-sensitive (color blindness, deutan) |
| chr5_-_74863514 | 0.01 |
ENSMUST00000117388.8
|
Lnx1
|
ligand of numb-protein X 1 |
| chr4_+_41903610 | 0.01 |
ENSMUST00000098128.4
|
Ccl21d
|
chemokine (C-C motif) ligand 21D |
| chr6_+_133269179 | 0.01 |
ENSMUST00000048459.8
|
5530400C23Rik
|
RIKEN cDNA 5530400C23 gene |
| chr14_+_64341735 | 0.01 |
ENSMUST00000022537.6
|
Prss52
|
protease, serine 52 |
| chr6_+_24857967 | 0.01 |
ENSMUST00000200968.4
|
Hyal5
|
hyaluronoglucosaminidase 5 |
| chr11_-_77784922 | 0.01 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr6_-_144155167 | 0.01 |
ENSMUST00000077160.12
|
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr2_-_111476340 | 0.01 |
ENSMUST00000119566.4
|
Olfr1298
|
olfactory receptor 1298 |
| chr10_+_129406526 | 0.01 |
ENSMUST00000059957.4
|
Olfr794
|
olfactory receptor 794 |
| chr16_-_22848153 | 0.01 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr3_+_132335575 | 0.01 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr15_-_57128522 | 0.01 |
ENSMUST00000137764.2
ENSMUST00000022995.13 |
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr2_-_85162315 | 0.01 |
ENSMUST00000099929.3
|
Olfr987
|
olfactory receptor 987 |
| chr10_-_128904846 | 0.01 |
ENSMUST00000204515.3
|
Olfr766-ps1
|
olfactory receptor 766, pseudogene 1 |
| chr7_+_17799849 | 0.01 |
ENSMUST00000032520.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr7_+_17799889 | 0.01 |
ENSMUST00000108483.2
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr12_-_113876103 | 0.01 |
ENSMUST00000193133.3
ENSMUST00000103462.4 |
Ighv7-2
|
immunoglobulin heavy variable 7-2 |
| chr5_-_89605622 | 0.01 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr7_-_139785877 | 0.01 |
ENSMUST00000215815.2
|
Olfr524
|
olfactory receptor 524 |
| chr2_-_111707494 | 0.01 |
ENSMUST00000213405.2
ENSMUST00000213737.2 |
Olfr1305
|
olfactory receptor 1305 |
| chr8_+_106154266 | 0.01 |
ENSMUST00000067305.8
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr2_+_90154541 | 0.01 |
ENSMUST00000077123.4
|
Olfr1565
|
olfactory receptor 1565 |
| chr6_-_57885013 | 0.01 |
ENSMUST00000228076.2
ENSMUST00000228905.2 ENSMUST00000227650.2 ENSMUST00000227342.2 ENSMUST00000228257.2 |
Vmn1r22
|
vomeronasal 1 receptor 22 |
| chr13_+_33508060 | 0.01 |
ENSMUST00000075515.7
|
Serpinb9f
|
serine (or cysteine) peptidase inhibitor, clade B, member 9f |
| chr7_-_28330322 | 0.01 |
ENSMUST00000040112.5
ENSMUST00000239470.2 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr9_+_38019424 | 0.01 |
ENSMUST00000211851.3
|
Olfr888
|
olfactory receptor 888 |
| chr5_-_137869969 | 0.01 |
ENSMUST00000196162.5
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chr4_+_3172083 | 0.01 |
ENSMUST00000105160.5
|
Vmn1r2
|
vomeronasal 1 receptor 2 |
| chr7_+_99384352 | 0.01 |
ENSMUST00000098264.2
|
Olfr520
|
olfactory receptor 520 |
| chr9_-_36040075 | 0.01 |
ENSMUST00000168452.2
|
Gm5916
|
predicted gene 5916 |
| chr10_-_62198043 | 0.01 |
ENSMUST00000149534.2
|
Hk1
|
hexokinase 1 |
| chr3_-_152931392 | 0.01 |
ENSMUST00000199707.2
|
St6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr16_-_22847760 | 0.01 |
ENSMUST00000039338.13
|
Kng2
|
kininogen 2 |
| chr4_-_156285247 | 0.01 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr6_+_57047880 | 0.01 |
ENSMUST00000171186.2
|
Vmn1r9
|
vomeronasal 1 receptor 9 |
| chr18_+_80298441 | 0.01 |
ENSMUST00000123750.8
|
Slc66a2
|
solute carrier family 66 member 2 |
| chr5_-_95419447 | 0.01 |
ENSMUST00000180232.2
|
Pramel46
|
PRAME like 46 |
| chr6_-_23248361 | 0.01 |
ENSMUST00000031709.7
|
Fezf1
|
Fez family zinc finger 1 |
| chr12_+_85017671 | 0.01 |
ENSMUST00000021669.15
ENSMUST00000171040.2 |
Fcf1
|
FCF1 rRNA processing protein |
| chr10_-_33662700 | 0.01 |
ENSMUST00000223295.2
|
Sult3a2
|
sulfotransferase family 3A, member 2 |
| chrX_+_148573187 | 0.01 |
ENSMUST00000112741.4
|
Gm15097
|
predicted gene 15097 |
| chr7_-_12370061 | 0.01 |
ENSMUST00000086210.6
|
Vmn2r54
|
vomeronasal 2, receptor 54 |
| chr9_-_35429565 | 0.01 |
ENSMUST00000183573.5
|
Gm1113
|
predicted gene 1113 |
| chr1_-_162694076 | 0.00 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr3_+_94172994 | 0.00 |
ENSMUST00000098878.4
|
Spopfm1
|
speckle-type BTB/POZ protein family member 1 |
| chr14_+_53913598 | 0.00 |
ENSMUST00000103662.6
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox17
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.3 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.1 | 0.3 | GO:0070859 | lipid transport involved in lipid storage(GO:0010877) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.0 | 0.1 | GO:0045914 | regulation of sulfur amino acid metabolic process(GO:0031335) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.3 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |