Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
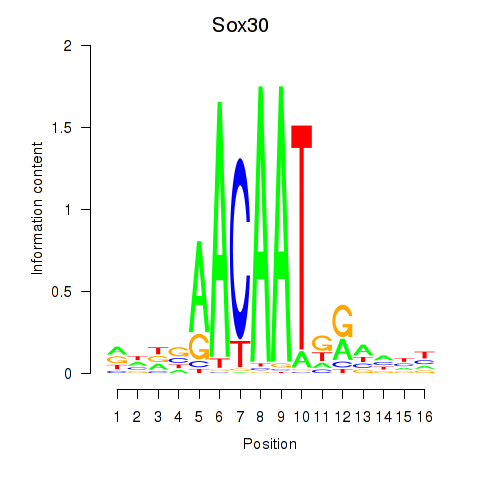
Results for Sox30
Z-value: 0.83
Transcription factors associated with Sox30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox30
|
ENSMUSG00000040489.6 | SRY (sex determining region Y)-box 30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox30 | mm39_v1_chr11_+_45871135_45871164 | 0.98 | 2.8e-03 | Click! |
Activity profile of Sox30 motif
Sorted Z-values of Sox30 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_133062677 | 0.77 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr3_-_102871440 | 0.61 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr9_+_20193647 | 0.61 |
ENSMUST00000071725.4
ENSMUST00000212983.3 |
Olfr39
|
olfactory receptor 39 |
| chr5_+_146769700 | 0.59 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chr19_-_24454720 | 0.52 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A |
| chr5_-_136275407 | 0.51 |
ENSMUST00000196245.2
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr2_+_85545763 | 0.40 |
ENSMUST00000216443.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr1_-_176641607 | 0.38 |
ENSMUST00000195717.6
ENSMUST00000192961.6 |
Cep170
|
centrosomal protein 170 |
| chr1_+_176642226 | 0.38 |
ENSMUST00000056773.15
ENSMUST00000027785.15 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr7_-_12853779 | 0.38 |
ENSMUST00000227220.2
ENSMUST00000227700.2 ENSMUST00000226604.2 |
Vmn1r86
|
vomeronasal 1 receptor 86 |
| chr4_+_83335947 | 0.38 |
ENSMUST00000030206.10
ENSMUST00000071544.11 |
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr9_-_32452885 | 0.35 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr6_-_87312743 | 0.35 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr1_+_178146689 | 0.34 |
ENSMUST00000027781.7
|
Cox20
|
cytochrome c oxidase assembly protein 20 |
| chr1_-_78488795 | 0.31 |
ENSMUST00000170511.3
|
BC035947
|
cDNA sequence BC035947 |
| chr4_-_119047167 | 0.30 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_119047202 | 0.29 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr11_-_49005701 | 0.29 |
ENSMUST00000060398.3
ENSMUST00000215553.2 ENSMUST00000109201.2 |
Olfr1396
|
olfactory receptor 1396 |
| chr5_+_14564932 | 0.28 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr8_-_23143422 | 0.28 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta |
| chr6_-_116084810 | 0.28 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr4_+_152160713 | 0.27 |
ENSMUST00000239025.2
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr11_-_58521327 | 0.26 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr10_-_62628008 | 0.25 |
ENSMUST00000217768.2
ENSMUST00000020268.7 ENSMUST00000218946.2 ENSMUST00000219527.2 |
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr6_-_122317156 | 0.25 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr8_+_109429732 | 0.25 |
ENSMUST00000188994.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr4_-_114991478 | 0.24 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr3_-_9898676 | 0.19 |
ENSMUST00000108384.9
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr11_-_99134885 | 0.18 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr3_-_9898713 | 0.17 |
ENSMUST00000161949.8
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr2_+_102488985 | 0.16 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr10_-_24712034 | 0.15 |
ENSMUST00000218044.2
ENSMUST00000020169.9 |
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr13_-_93774469 | 0.13 |
ENSMUST00000099309.6
|
Bhmt
|
betaine-homocysteine methyltransferase |
| chr14_-_19873839 | 0.11 |
ENSMUST00000022341.7
|
Rtraf
|
RNA transcription, translation and transport factor |
| chr19_-_56810593 | 0.10 |
ENSMUST00000118592.8
|
Ccdc186
|
coiled-coil domain containing 186 |
| chr6_-_132291714 | 0.08 |
ENSMUST00000048686.9
|
Prpmp5
|
proline-rich protein MP5 |
| chr18_+_37610858 | 0.08 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr9_+_74883377 | 0.08 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr17_-_79662514 | 0.08 |
ENSMUST00000068958.9
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr9_-_72018453 | 0.08 |
ENSMUST00000183992.8
|
Tcf12
|
transcription factor 12 |
| chr1_-_151376544 | 0.07 |
ENSMUST00000187991.2
ENSMUST00000187048.7 ENSMUST00000186415.7 |
Rnf2
|
ring finger protein 2 |
| chr11_+_21041291 | 0.07 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr9_+_7764042 | 0.07 |
ENSMUST00000052865.16
|
Tmem123
|
transmembrane protein 123 |
| chr4_+_109200225 | 0.06 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr9_-_60418286 | 0.06 |
ENSMUST00000098660.10
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr3_-_113198765 | 0.05 |
ENSMUST00000179568.3
|
Amy2a4
|
amylase 2a4 |
| chr6_-_12749192 | 0.05 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr2_-_57942844 | 0.05 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr6_-_136899167 | 0.04 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr1_-_37535170 | 0.04 |
ENSMUST00000148047.2
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr5_-_137101108 | 0.04 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr9_-_72019053 | 0.04 |
ENSMUST00000183492.8
ENSMUST00000184523.8 ENSMUST00000034755.13 |
Tcf12
|
transcription factor 12 |
| chr2_+_69869459 | 0.04 |
ENSMUST00000060208.11
|
Myo3b
|
myosin IIIB |
| chr9_-_19968858 | 0.03 |
ENSMUST00000216538.2
ENSMUST00000212098.3 |
Olfr867
|
olfactory receptor 867 |
| chr7_+_119499322 | 0.03 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr11_-_116572116 | 0.03 |
ENSMUST00000144398.2
|
St6galnac2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr5_+_139197689 | 0.03 |
ENSMUST00000148772.8
ENSMUST00000110882.8 |
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr7_+_43079512 | 0.02 |
ENSMUST00000004732.7
|
Lim2
|
lens intrinsic membrane protein 2 |
| chr2_-_65397850 | 0.02 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr5_+_102992873 | 0.02 |
ENSMUST00000070000.6
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr7_+_43874752 | 0.02 |
ENSMUST00000075162.5
|
Klk1
|
kallikrein 1 |
| chr18_-_68562385 | 0.02 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr17_-_67354971 | 0.01 |
ENSMUST00000224862.2
|
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chr11_+_43365103 | 0.01 |
ENSMUST00000173002.8
ENSMUST00000057679.10 |
C1qtnf2
|
C1q and tumor necrosis factor related protein 2 |
| chr2_-_84652890 | 0.01 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr1_+_38037086 | 0.01 |
ENSMUST00000027252.8
|
Eif5b
|
eukaryotic translation initiation factor 5B |
| chr17_-_34337446 | 0.01 |
ENSMUST00000095347.13
|
Brd2
|
bromodomain containing 2 |
| chr1_+_78488499 | 0.01 |
ENSMUST00000012331.7
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr17_+_31605184 | 0.01 |
ENSMUST00000047168.13
ENSMUST00000127929.8 ENSMUST00000134525.9 ENSMUST00000236454.2 ENSMUST00000238091.2 ENSMUST00000235719.2 |
Pde9a
|
phosphodiesterase 9A |
| chr18_+_37875135 | 0.00 |
ENSMUST00000003599.9
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr2_-_35257741 | 0.00 |
ENSMUST00000028243.2
|
4930568D16Rik
|
RIKEN cDNA 4930568D16 gene |
| chr1_+_78488440 | 0.00 |
ENSMUST00000152111.2
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr7_+_43661592 | 0.00 |
ENSMUST00000048945.6
|
Klk1b26
|
kallikrein 1-related petidase b26 |
| chrX_-_93408176 | 0.00 |
ENSMUST00000239046.2
|
CXorf58
|
family with sequence similarity 90, member A1B |
| chr4_+_48585275 | 0.00 |
ENSMUST00000123476.8
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr2_+_3115250 | 0.00 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr2_-_101713355 | 0.00 |
ENSMUST00000154525.2
|
Prr5l
|
proline rich 5 like |
| chr4_+_48585135 | 0.00 |
ENSMUST00000030032.13
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox30
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 0.3 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.5 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.4 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |