Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
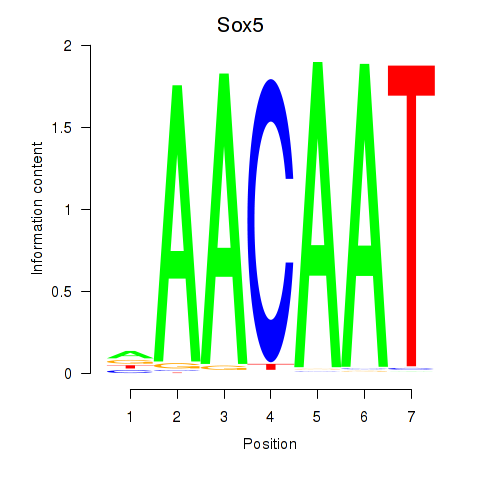
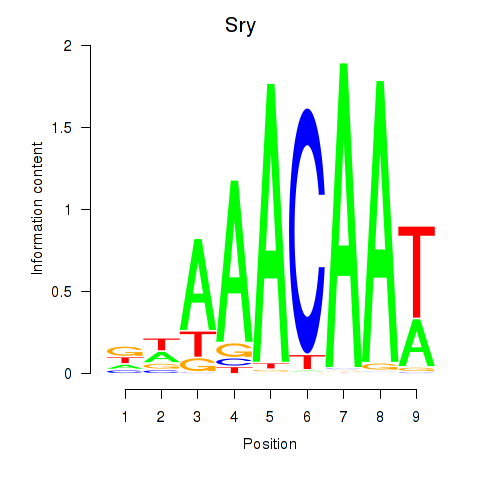
Results for Sox5_Sry
Z-value: 0.51
Transcription factors associated with Sox5_Sry
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox5
|
ENSMUSG00000041540.17 | SRY (sex determining region Y)-box 5 |
|
Sry
|
ENSMUSG00000069036.4 | sex determining region of Chr Y |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox5 | mm39_v1_chr6_-_144155167_144155195 | 0.72 | 1.7e-01 | Click! |
| Sry | mm39_v1_chrY_-_2663658_2663658 | 0.38 | 5.2e-01 | Click! |
Activity profile of Sox5_Sry motif
Sorted Z-values of Sox5_Sry motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_50183129 | 0.57 |
ENSMUST00000059458.5
|
Maml1
|
mastermind like transcriptional coactivator 1 |
| chr16_-_4031814 | 0.47 |
ENSMUST00000023165.9
|
Crebbp
|
CREB binding protein |
| chr1_+_34044940 | 0.42 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr9_-_49710058 | 0.40 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr5_-_66238313 | 0.35 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr6_+_15185202 | 0.33 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr9_-_49710190 | 0.31 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr12_+_40495951 | 0.27 |
ENSMUST00000037488.8
|
Dock4
|
dedicator of cytokinesis 4 |
| chr1_+_143516402 | 0.27 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr9_+_119978773 | 0.25 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr5_+_75316552 | 0.25 |
ENSMUST00000168162.5
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr2_-_168443540 | 0.24 |
ENSMUST00000171689.8
ENSMUST00000137451.2 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr4_-_128699838 | 0.23 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chr12_-_99359265 | 0.23 |
ENSMUST00000177451.8
|
Foxn3
|
forkhead box N3 |
| chr5_-_142803135 | 0.22 |
ENSMUST00000198181.2
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr13_+_83720484 | 0.21 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chrM_-_14061 | 0.20 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr9_-_48747232 | 0.20 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr8_+_66419809 | 0.19 |
ENSMUST00000072482.13
|
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr13_+_83720457 | 0.19 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr1_+_39940189 | 0.19 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr2_-_104324035 | 0.19 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr16_+_57369595 | 0.18 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr6_-_87312743 | 0.18 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr7_+_79460475 | 0.18 |
ENSMUST00000107394.3
|
Mesp2
|
mesoderm posterior 2 |
| chr9_-_32452885 | 0.18 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr9_+_27702243 | 0.17 |
ENSMUST00000115243.9
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr5_+_42225303 | 0.17 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr10_-_45346297 | 0.17 |
ENSMUST00000079390.7
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr19_-_4384029 | 0.17 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr4_+_122889737 | 0.16 |
ENSMUST00000106252.9
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr10_+_17598961 | 0.16 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr8_+_3543131 | 0.14 |
ENSMUST00000061508.8
ENSMUST00000207318.2 |
Zfp358
|
zinc finger protein 358 |
| chr16_-_4698148 | 0.14 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr11_-_110142565 | 0.14 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr16_-_89940652 | 0.14 |
ENSMUST00000114124.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr4_+_129229373 | 0.14 |
ENSMUST00000141235.8
|
Zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chrX_+_158623460 | 0.14 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr4_+_122889828 | 0.13 |
ENSMUST00000030407.8
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr4_+_137720326 | 0.13 |
ENSMUST00000139759.8
ENSMUST00000058133.10 ENSMUST00000105830.9 ENSMUST00000084215.12 |
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr6_-_120341304 | 0.13 |
ENSMUST00000146667.2
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr14_-_100521888 | 0.13 |
ENSMUST00000226774.2
|
Klf12
|
Kruppel-like factor 12 |
| chr9_-_32454157 | 0.13 |
ENSMUST00000183767.2
|
Fli1
|
Friend leukemia integration 1 |
| chr13_-_40887244 | 0.13 |
ENSMUST00000110193.9
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr3_+_101284391 | 0.13 |
ENSMUST00000043983.11
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr2_-_101627999 | 0.12 |
ENSMUST00000171088.8
ENSMUST00000043845.14 |
Prr5l
|
proline rich 5 like |
| chr6_+_122803624 | 0.12 |
ENSMUST00000203075.2
|
Foxj2
|
forkhead box J2 |
| chr7_+_96730915 | 0.12 |
ENSMUST00000206791.2
|
Gab2
|
growth factor receptor bound protein 2-associated protein 2 |
| chr18_+_35695736 | 0.12 |
ENSMUST00000235851.2
ENSMUST00000235581.2 |
Matr3
|
matrin 3 |
| chr6_+_17491231 | 0.12 |
ENSMUST00000080469.12
|
Met
|
met proto-oncogene |
| chr6_-_149003003 | 0.12 |
ENSMUST00000127727.2
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr17_+_47747657 | 0.12 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr7_+_126359869 | 0.11 |
ENSMUST00000206272.2
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr4_+_83335947 | 0.11 |
ENSMUST00000030206.10
ENSMUST00000071544.11 |
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr6_+_136509922 | 0.11 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr19_-_31742427 | 0.11 |
ENSMUST00000065067.14
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr9_+_20193647 | 0.11 |
ENSMUST00000071725.4
ENSMUST00000212983.3 |
Olfr39
|
olfactory receptor 39 |
| chr8_-_107064615 | 0.10 |
ENSMUST00000067512.8
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr1_-_84817976 | 0.10 |
ENSMUST00000190067.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr3_-_95725944 | 0.10 |
ENSMUST00000200164.5
ENSMUST00000090791.8 ENSMUST00000197449.2 |
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr2_+_92015780 | 0.10 |
ENSMUST00000128781.9
ENSMUST00000111291.9 |
Phf21a
|
PHD finger protein 21A |
| chr3_+_63203235 | 0.10 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr1_+_59952131 | 0.10 |
ENSMUST00000036540.12
|
Fam117b
|
family with sequence similarity 117, member B |
| chr10_-_68114543 | 0.10 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr4_-_33248499 | 0.10 |
ENSMUST00000049357.10
|
Pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr8_+_109429732 | 0.10 |
ENSMUST00000188994.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr1_-_167112170 | 0.09 |
ENSMUST00000192269.3
|
Uck2
|
uridine-cytidine kinase 2 |
| chr11_+_60244132 | 0.09 |
ENSMUST00000070805.13
ENSMUST00000094140.9 ENSMUST00000108723.9 ENSMUST00000108722.5 |
Drc3
|
dynein regulatory complex subunit 3 |
| chr18_-_15196612 | 0.09 |
ENSMUST00000168989.9
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr19_+_23736205 | 0.09 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr11_-_103254257 | 0.09 |
ENSMUST00000092557.6
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr8_-_84963653 | 0.09 |
ENSMUST00000039480.7
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr1_+_176642226 | 0.09 |
ENSMUST00000056773.15
ENSMUST00000027785.15 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr8_-_85500998 | 0.09 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr1_-_176641607 | 0.09 |
ENSMUST00000195717.6
ENSMUST00000192961.6 |
Cep170
|
centrosomal protein 170 |
| chr19_-_24454720 | 0.09 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A |
| chr13_+_83723743 | 0.09 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr14_-_45456821 | 0.09 |
ENSMUST00000128484.8
ENSMUST00000147853.8 ENSMUST00000022377.11 ENSMUST00000143609.2 ENSMUST00000153383.8 ENSMUST00000139526.9 |
Txndc16
|
thioredoxin domain containing 16 |
| chr11_-_101917745 | 0.09 |
ENSMUST00000107167.2
ENSMUST00000062801.11 |
Mpp3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr11_-_58353096 | 0.09 |
ENSMUST00000215691.2
|
Olfr30
|
olfactory receptor 30 |
| chr2_+_18059982 | 0.09 |
ENSMUST00000028076.15
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr4_+_32238712 | 0.09 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr7_+_127475968 | 0.09 |
ENSMUST00000131000.2
|
Zfp646
|
zinc finger protein 646 |
| chr12_-_40495753 | 0.09 |
ENSMUST00000069692.10
ENSMUST00000069637.15 |
Zfp277
|
zinc finger protein 277 |
| chr10_+_87694924 | 0.08 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr3_+_90161470 | 0.08 |
ENSMUST00000029545.15
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr13_-_115238427 | 0.08 |
ENSMUST00000224997.2
ENSMUST00000061673.9 |
Gm49395
Itga1
|
predicted gene, 49395 integrin alpha 1 |
| chr14_+_26359390 | 0.08 |
ENSMUST00000112318.10
|
Arf4
|
ADP-ribosylation factor 4 |
| chr1_-_80687213 | 0.08 |
ENSMUST00000186087.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr13_+_38010879 | 0.08 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr4_+_152160713 | 0.08 |
ENSMUST00000239025.2
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chrX_-_142716200 | 0.08 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr2_+_146063841 | 0.08 |
ENSMUST00000089257.6
|
Insm1
|
insulinoma-associated 1 |
| chr17_-_10501816 | 0.08 |
ENSMUST00000233684.2
|
Qk
|
quaking, KH domain containing RNA binding |
| chr13_+_38009981 | 0.08 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr9_+_44398524 | 0.08 |
ENSMUST00000218183.2
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr2_+_102488985 | 0.08 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr16_-_16377982 | 0.08 |
ENSMUST00000161861.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr3_+_53396120 | 0.08 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr9_-_62888156 | 0.08 |
ENSMUST00000098651.6
ENSMUST00000214830.2 |
Pias1
|
protein inhibitor of activated STAT 1 |
| chr6_-_116084810 | 0.07 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr9_-_102231884 | 0.07 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chr7_+_16044336 | 0.07 |
ENSMUST00000136781.2
|
Bbc3
|
BCL2 binding component 3 |
| chr9_+_117869642 | 0.07 |
ENSMUST00000134433.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr11_-_86884507 | 0.07 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chrX_+_41238410 | 0.07 |
ENSMUST00000127618.8
|
Stag2
|
stromal antigen 2 |
| chr2_+_154278357 | 0.07 |
ENSMUST00000109725.8
ENSMUST00000099178.10 ENSMUST00000045270.15 ENSMUST00000109724.2 |
Cbfa2t2
|
CBFA2/RUNX1 translocation partner 2 |
| chr11_+_11635908 | 0.07 |
ENSMUST00000065433.12
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr5_-_123859070 | 0.07 |
ENSMUST00000031376.12
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr15_-_80929669 | 0.07 |
ENSMUST00000135047.2
|
Mrtfa
|
myocardin related transcription factor A |
| chr5_-_86666408 | 0.07 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr12_-_76224025 | 0.07 |
ENSMUST00000101291.11
ENSMUST00000218621.2 ENSMUST00000076634.5 |
Esr2
|
estrogen receptor 2 (beta) |
| chr2_-_59955995 | 0.07 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr5_+_14564932 | 0.07 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr8_+_65399831 | 0.07 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr15_-_78004211 | 0.07 |
ENSMUST00000019290.3
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr4_-_103072343 | 0.07 |
ENSMUST00000150285.8
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr13_+_41040657 | 0.07 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr1_+_39940043 | 0.07 |
ENSMUST00000168431.7
ENSMUST00000163854.9 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr12_-_81827893 | 0.07 |
ENSMUST00000035987.9
|
Map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr2_-_57014015 | 0.06 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_-_125839897 | 0.06 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr2_-_63928339 | 0.06 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr11_+_11636213 | 0.06 |
ENSMUST00000076700.11
ENSMUST00000048122.13 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr15_+_99600475 | 0.06 |
ENSMUST00000228984.2
ENSMUST00000229845.2 |
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr7_-_28071658 | 0.06 |
ENSMUST00000094644.11
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr6_-_122317156 | 0.06 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr6_+_120341055 | 0.06 |
ENSMUST00000005108.10
|
Kdm5a
|
lysine (K)-specific demethylase 5A |
| chr15_-_50746202 | 0.06 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr18_-_16942289 | 0.06 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr4_-_135601098 | 0.06 |
ENSMUST00000030436.12
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chrX_-_72868544 | 0.06 |
ENSMUST00000002080.12
ENSMUST00000114438.3 |
Pdzd4
|
PDZ domain containing 4 |
| chr14_-_71004019 | 0.06 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr3_-_96833336 | 0.06 |
ENSMUST00000062944.7
|
Gja8
|
gap junction protein, alpha 8 |
| chr8_-_85500010 | 0.06 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr7_+_128290204 | 0.06 |
ENSMUST00000118605.2
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr12_-_72283465 | 0.06 |
ENSMUST00000021497.16
ENSMUST00000137990.2 |
Rtn1
|
reticulon 1 |
| chr17_+_47451868 | 0.06 |
ENSMUST00000190080.9
|
Trerf1
|
transcriptional regulating factor 1 |
| chr4_-_70453140 | 0.06 |
ENSMUST00000107359.9
|
Megf9
|
multiple EGF-like-domains 9 |
| chr9_+_117869543 | 0.06 |
ENSMUST00000044454.12
|
Azi2
|
5-azacytidine induced gene 2 |
| chr5_+_113873864 | 0.06 |
ENSMUST00000065698.7
|
Ficd
|
FIC domain containing |
| chr10_-_62628008 | 0.06 |
ENSMUST00000217768.2
ENSMUST00000020268.7 ENSMUST00000218946.2 ENSMUST00000219527.2 |
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr2_+_112114906 | 0.06 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr15_+_25622611 | 0.05 |
ENSMUST00000110457.8
ENSMUST00000137601.8 |
Myo10
|
myosin X |
| chr19_+_26600820 | 0.05 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_+_6423651 | 0.05 |
ENSMUST00000125516.8
|
Sf1
|
splicing factor 1 |
| chr6_-_56774622 | 0.05 |
ENSMUST00000114323.8
|
Kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr5_-_131645437 | 0.05 |
ENSMUST00000161804.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr1_-_136273436 | 0.05 |
ENSMUST00000192001.6
ENSMUST00000192314.2 |
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr9_+_117869577 | 0.05 |
ENSMUST00000133580.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr18_-_60881679 | 0.05 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr7_+_75105282 | 0.05 |
ENSMUST00000207750.2
ENSMUST00000166315.7 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr16_-_95260104 | 0.05 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr1_-_57011595 | 0.05 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr5_+_34140777 | 0.05 |
ENSMUST00000094869.12
ENSMUST00000114383.8 |
Gm1673
|
predicted gene 1673 |
| chr2_-_35994072 | 0.05 |
ENSMUST00000112961.10
ENSMUST00000112966.10 |
Lhx6
|
LIM homeobox protein 6 |
| chr12_-_32000534 | 0.05 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr9_+_61279640 | 0.05 |
ENSMUST00000178113.8
ENSMUST00000159386.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr15_+_99600149 | 0.05 |
ENSMUST00000229236.2
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr11_-_58521327 | 0.05 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr15_+_101164719 | 0.05 |
ENSMUST00000230814.2
ENSMUST00000023779.8 |
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr7_+_79675727 | 0.05 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr14_-_57983511 | 0.05 |
ENSMUST00000173990.8
ENSMUST00000022531.14 |
Lats2
|
large tumor suppressor 2 |
| chrX_+_56008685 | 0.05 |
ENSMUST00000096431.10
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr8_+_121215155 | 0.05 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr9_-_72018453 | 0.05 |
ENSMUST00000183992.8
|
Tcf12
|
transcription factor 12 |
| chr14_+_45457168 | 0.05 |
ENSMUST00000227086.2
ENSMUST00000147957.2 |
Gpr137c
|
G protein-coupled receptor 137C |
| chr17_-_30107544 | 0.05 |
ENSMUST00000171691.9
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr18_+_69726654 | 0.05 |
ENSMUST00000200921.4
|
Tcf4
|
transcription factor 4 |
| chr11_-_58221145 | 0.05 |
ENSMUST00000186859.2
ENSMUST00000065533.3 |
Zfp672
|
zinc finger protein 672 |
| chrX_+_41239872 | 0.04 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2 |
| chr2_+_32425327 | 0.04 |
ENSMUST00000133512.2
ENSMUST00000048375.6 |
Fam102a
|
family with sequence similarity 102, member A |
| chr5_-_123859153 | 0.04 |
ENSMUST00000196282.5
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr11_+_96162283 | 0.04 |
ENSMUST00000000010.9
ENSMUST00000174042.3 |
Hoxb9
|
homeobox B9 |
| chr10_-_8638215 | 0.04 |
ENSMUST00000212553.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr19_+_53317844 | 0.04 |
ENSMUST00000111737.3
ENSMUST00000025998.15 ENSMUST00000237837.2 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr1_-_64160557 | 0.04 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_+_44508137 | 0.04 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chr6_-_87312681 | 0.04 |
ENSMUST00000204805.3
|
Antxr1
|
anthrax toxin receptor 1 |
| chr5_-_86521273 | 0.04 |
ENSMUST00000031175.12
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr4_+_109200225 | 0.04 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr18_+_65022035 | 0.04 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr9_-_63929308 | 0.04 |
ENSMUST00000179458.2
ENSMUST00000041029.6 |
Smad6
|
SMAD family member 6 |
| chr11_-_115704447 | 0.04 |
ENSMUST00000041684.11
ENSMUST00000156812.2 |
Caskin2
|
CASK-interacting protein 2 |
| chr11_-_60243695 | 0.04 |
ENSMUST00000095254.12
ENSMUST00000102683.11 ENSMUST00000093048.13 ENSMUST00000093046.13 ENSMUST00000064019.15 ENSMUST00000102682.5 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr5_-_136595205 | 0.04 |
ENSMUST00000176778.8
|
Cux1
|
cut-like homeobox 1 |
| chr8_+_121854566 | 0.04 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr19_+_38252984 | 0.04 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr11_-_51891259 | 0.04 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr1_-_119765068 | 0.04 |
ENSMUST00000163435.8
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr18_-_3337467 | 0.04 |
ENSMUST00000154135.8
ENSMUST00000142690.2 ENSMUST00000025069.11 ENSMUST00000165086.8 ENSMUST00000082141.12 ENSMUST00000149803.8 |
Crem
|
cAMP responsive element modulator |
| chr3_+_95067759 | 0.04 |
ENSMUST00000131742.8
ENSMUST00000090823.8 ENSMUST00000090821.10 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr2_-_172296662 | 0.04 |
ENSMUST00000161334.2
|
Gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr6_-_12749192 | 0.04 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr2_+_121001636 | 0.04 |
ENSMUST00000110658.8
|
Tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr6_-_52203146 | 0.03 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr9_+_110361561 | 0.03 |
ENSMUST00000153838.8
|
Setd2
|
SET domain containing 2 |
| chr2_-_118380149 | 0.03 |
ENSMUST00000090219.13
|
Bmf
|
BCL2 modifying factor |
| chr4_-_129015682 | 0.03 |
ENSMUST00000139450.8
ENSMUST00000125931.9 ENSMUST00000116444.10 |
Hpca
|
hippocalcin |
| chr6_-_99243455 | 0.03 |
ENSMUST00000113326.9
|
Foxp1
|
forkhead box P1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox5_Sry
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 0.6 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.5 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.1 | 0.2 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.0 | 0.1 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.0 | 0.5 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0060971 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.3 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.1 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.0 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.0 | 0.0 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 0.0 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.7 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |