Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
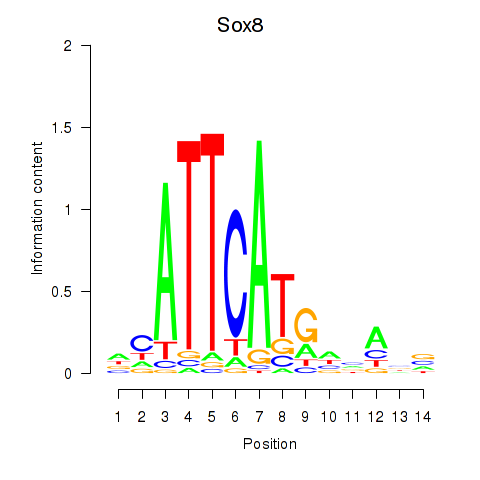
Results for Sox8
Z-value: 0.77
Transcription factors associated with Sox8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox8
|
ENSMUSG00000024176.11 | SRY (sex determining region Y)-box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox8 | mm39_v1_chr17_-_25789652_25789670 | -0.74 | 1.5e-01 | Click! |
Activity profile of Sox8 motif
Sorted Z-values of Sox8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_126399208 | 0.77 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_126399778 | 0.74 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_126399574 | 0.56 |
ENSMUST00000106348.8
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr4_-_133972890 | 0.47 |
ENSMUST00000030644.8
|
Zfp593
|
zinc finger protein 593 |
| chr13_-_19579898 | 0.40 |
ENSMUST00000197565.3
ENSMUST00000221380.2 ENSMUST00000200323.3 ENSMUST00000199924.2 ENSMUST00000222869.2 |
Stard3nl
|
STARD3 N-terminal like |
| chr10_+_11157326 | 0.37 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr13_-_19579961 | 0.32 |
ENSMUST00000039694.13
|
Stard3nl
|
STARD3 N-terminal like |
| chr14_+_21881794 | 0.25 |
ENSMUST00000152562.8
|
Vdac2
|
voltage-dependent anion channel 2 |
| chr11_-_43638790 | 0.24 |
ENSMUST00000048578.3
ENSMUST00000109278.8 |
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr4_+_154226819 | 0.21 |
ENSMUST00000030895.12
|
Wrap73
|
WD repeat containing, antisense to Trp73 |
| chr13_+_93441307 | 0.21 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr15_-_103123711 | 0.20 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chrX_-_71961890 | 0.20 |
ENSMUST00000152200.2
|
Cetn2
|
centrin 2 |
| chr6_-_72357398 | 0.20 |
ENSMUST00000101285.10
ENSMUST00000074231.6 |
Vamp5
|
vesicle-associated membrane protein 5 |
| chr10_+_58207229 | 0.19 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_-_26052335 | 0.17 |
ENSMUST00000044911.10
ENSMUST00000237186.2 |
Stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr13_-_31134281 | 0.17 |
ENSMUST00000102946.8
|
Exoc2
|
exocyst complex component 2 |
| chrX_+_157993303 | 0.15 |
ENSMUST00000112493.8
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr18_-_84969601 | 0.15 |
ENSMUST00000025547.4
|
Timm21
|
translocase of inner mitochondrial membrane 21 |
| chr4_+_11758147 | 0.15 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr13_+_93440572 | 0.14 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr2_+_4405769 | 0.13 |
ENSMUST00000075767.14
|
Frmd4a
|
FERM domain containing 4A |
| chr2_+_85809620 | 0.12 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr4_-_116263183 | 0.11 |
ENSMUST00000123072.8
ENSMUST00000144281.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr13_-_81859056 | 0.11 |
ENSMUST00000161920.2
ENSMUST00000048993.12 |
Polr3g
|
polymerase (RNA) III (DNA directed) polypeptide G |
| chr5_+_90666791 | 0.08 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr13_+_19528728 | 0.08 |
ENSMUST00000179181.3
|
Trgc4
|
T cell receptor gamma, constant 4 |
| chr1_-_79838897 | 0.07 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr5_+_53424471 | 0.07 |
ENSMUST00000147148.5
|
Smim20
|
small integral membrane protein 20 |
| chr9_+_108566513 | 0.07 |
ENSMUST00000192344.2
|
Prkar2a
|
protein kinase, cAMP dependent regulatory, type II alpha |
| chr2_+_109522781 | 0.07 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr9_+_109881083 | 0.05 |
ENSMUST00000164930.8
ENSMUST00000199498.5 |
Map4
|
microtubule-associated protein 4 |
| chr12_-_114955196 | 0.05 |
ENSMUST00000194865.2
|
Ighv1-47
|
immunoglobulin heavy variable 1-47 |
| chrX_+_133305529 | 0.04 |
ENSMUST00000113224.9
ENSMUST00000113226.2 |
Drp2
|
dystrophin related protein 2 |
| chr19_-_33764859 | 0.04 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr6_-_132688891 | 0.04 |
ENSMUST00000105077.2
|
Tas2r122
|
taste receptor, type 2, member 122 |
| chr5_-_44259010 | 0.04 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr5_-_44259293 | 0.04 |
ENSMUST00000074113.13
|
Prom1
|
prominin 1 |
| chr1_-_157072060 | 0.03 |
ENSMUST00000129880.8
|
Rasal2
|
RAS protein activator like 2 |
| chr1_+_183199384 | 0.03 |
ENSMUST00000065900.10
|
Hhipl2
|
hedgehog interacting protein-like 2 |
| chr12_-_115722081 | 0.03 |
ENSMUST00000103541.3
|
Ighv1-72
|
immunoglobulin heavy variable 1-72 |
| chr12_-_115083839 | 0.03 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr11_-_109986763 | 0.02 |
ENSMUST00000046223.14
ENSMUST00000106664.10 ENSMUST00000106662.2 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr12_-_115172211 | 0.02 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr2_-_85245221 | 0.02 |
ENSMUST00000099926.2
|
Olfr993
|
olfactory receptor 993 |
| chr16_-_58906844 | 0.02 |
ENSMUST00000078517.2
|
Olfr191
|
olfactory receptor 191 |
| chr11_-_99884818 | 0.02 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr19_+_13745671 | 0.02 |
ENSMUST00000061669.3
|
Olfr1495
|
olfactory receptor 1495 |
| chr14_-_52648384 | 0.02 |
ENSMUST00000079459.4
|
Olfr1510
|
olfactory receptor 1510 |
| chr7_+_106513407 | 0.02 |
ENSMUST00000098140.2
|
Olfr1532-ps1
|
olfactory receptor 1532, pseudogene 1 |
| chr10_-_129678931 | 0.02 |
ENSMUST00000057775.3
|
Olfr812
|
olfactory receptor 812 |
| chr18_+_37637317 | 0.01 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr17_-_40553176 | 0.01 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr6_-_42524521 | 0.01 |
ENSMUST00000217978.2
|
Olfr455
|
olfactory receptor 455 |
| chr11_-_109986804 | 0.01 |
ENSMUST00000100287.9
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr2_-_85230875 | 0.01 |
ENSMUST00000099927.2
|
Olfr992
|
olfactory receptor 992 |
| chr13_+_27241551 | 0.01 |
ENSMUST00000110369.10
ENSMUST00000224228.2 ENSMUST00000018061.7 |
Prl
|
prolactin |
| chr12_-_115425105 | 0.01 |
ENSMUST00000103532.3
|
Ighv1-62-3
|
immunoglobulin heavy variable 1-62-3 |
| chr14_-_8457069 | 0.01 |
ENSMUST00000022257.4
|
Atxn7
|
ataxin 7 |
| chr7_-_47677345 | 0.01 |
ENSMUST00000094390.3
|
Mrgprx1
|
MAS-related GPR, member X1 |
| chr1_+_109911467 | 0.01 |
ENSMUST00000172005.8
|
Cdh7
|
cadherin 7, type 2 |
| chr15_-_96918203 | 0.01 |
ENSMUST00000166223.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr11_-_99241924 | 0.01 |
ENSMUST00000017732.3
|
Krt27
|
keratin 27 |
| chr12_+_78243846 | 0.01 |
ENSMUST00000188791.2
|
Gm6657
|
predicted gene 6657 |
| chr13_-_21327251 | 0.01 |
ENSMUST00000055298.6
|
Olfr1368
|
olfactory receptor 1368 |
| chr4_-_58553553 | 0.01 |
ENSMUST00000107575.9
ENSMUST00000107574.8 ENSMUST00000147354.8 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr19_+_4771089 | 0.01 |
ENSMUST00000238976.3
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr7_+_108021780 | 0.01 |
ENSMUST00000076406.2
|
Olfr497
|
olfactory receptor 497 |
| chr7_+_102423702 | 0.01 |
ENSMUST00000098217.3
|
Olfr561
|
olfactory receptor 561 |
| chr6_+_115908709 | 0.01 |
ENSMUST00000032471.9
|
Rho
|
rhodopsin |
| chr9_-_95697441 | 0.01 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr11_+_69882410 | 0.01 |
ENSMUST00000108592.2
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr7_-_86035866 | 0.01 |
ENSMUST00000077210.3
|
Olfr304
|
olfactory receptor 304 |
| chr16_+_65320163 | 0.00 |
ENSMUST00000184525.2
|
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr19_+_13717092 | 0.00 |
ENSMUST00000207836.3
|
Olfr1494
|
olfactory receptor 1494 |
| chr1_+_135710803 | 0.00 |
ENSMUST00000132795.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr2_+_116709179 | 0.00 |
ENSMUST00000028834.3
|
Tmco5
|
transmembrane and coiled-coil domains 5 |
| chr7_-_106526664 | 0.00 |
ENSMUST00000072368.3
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr2_-_73605387 | 0.00 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr5_+_43673093 | 0.00 |
ENSMUST00000144558.3
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr2_+_116709167 | 0.00 |
ENSMUST00000123598.8
ENSMUST00000155470.8 |
Tmco5
|
transmembrane and coiled-coil domains 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.5 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.2 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |