Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
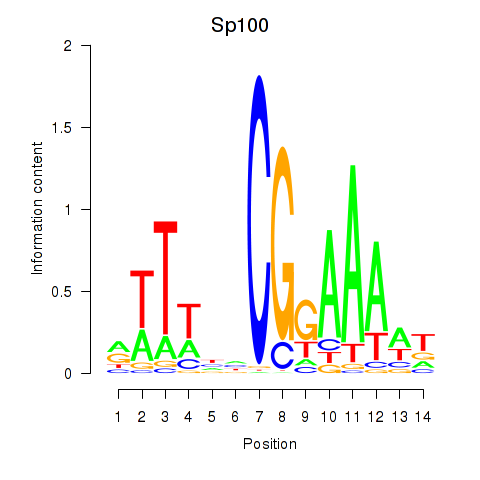
Results for Sp100
Z-value: 1.32
Transcription factors associated with Sp100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp100
|
ENSMUSG00000026222.17 | nuclear antigen Sp100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp100 | mm39_v1_chr1_+_85577766_85577774 | 0.72 | 1.7e-01 | Click! |
Activity profile of Sp100 motif
Sorted Z-values of Sp100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_23946359 | 4.15 |
ENSMUST00000091701.3
|
H3c1
|
H3 clustered histone 1 |
| chr9_+_3023547 | 2.13 |
ENSMUST00000099046.4
|
Gm10718
|
predicted gene 10718 |
| chr9_+_3013140 | 1.97 |
ENSMUST00000143083.3
|
Gm10721
|
predicted gene 10721 |
| chr9_+_3025417 | 1.66 |
ENSMUST00000075573.7
|
Gm10717
|
predicted gene 10717 |
| chr9_+_3018753 | 1.59 |
ENSMUST00000179272.2
|
Gm10719
|
predicted gene 10719 |
| chr9_+_3017408 | 1.50 |
ENSMUST00000099049.4
|
Gm10719
|
predicted gene 10719 |
| chr9_+_3004457 | 1.27 |
ENSMUST00000178348.2
|
Gm11168
|
predicted gene 11168 |
| chr13_+_23930717 | 1.25 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr9_+_3000922 | 1.15 |
ENSMUST00000151376.3
|
Gm10722
|
predicted gene 10722 |
| chr9_+_3027439 | 1.06 |
ENSMUST00000179982.2
ENSMUST00000177875.8 |
Gm10717
|
predicted gene 10717 |
| chr9_+_3032001 | 0.90 |
ENSMUST00000099056.4
ENSMUST00000179839.8 |
Gm10717
|
predicted gene 10717 |
| chr2_-_98497609 | 0.87 |
ENSMUST00000099683.2
|
Gm10800
|
predicted gene 10800 |
| chr18_+_61178211 | 0.85 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr7_-_3551003 | 0.76 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr9_+_3036877 | 0.71 |
ENSMUST00000155807.3
|
Gm10715
|
predicted gene 10715 |
| chr9_+_3015654 | 0.60 |
ENSMUST00000099050.4
|
Gm10720
|
predicted gene 10720 |
| chr5_+_16758538 | 0.57 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor |
| chrM_+_9870 | 0.57 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr15_-_5273659 | 0.57 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr15_-_5273645 | 0.55 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr2_+_128971620 | 0.52 |
ENSMUST00000035481.5
|
Chchd5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr7_+_4795873 | 0.46 |
ENSMUST00000032597.12
ENSMUST00000078432.5 |
Rpl28
|
ribosomal protein L28 |
| chr5_+_16758777 | 0.46 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr11_-_16458484 | 0.41 |
ENSMUST00000109643.8
ENSMUST00000166950.8 ENSMUST00000178855.8 |
Sec61g
|
SEC61, gamma subunit |
| chr7_-_3723381 | 0.39 |
ENSMUST00000078451.7
|
Pirb
|
paired Ig-like receptor B |
| chr11_-_16458069 | 0.38 |
ENSMUST00000109641.2
|
Sec61g
|
SEC61, gamma subunit |
| chr5_-_137015683 | 0.37 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr11_-_16458148 | 0.37 |
ENSMUST00000109642.8
|
Sec61g
|
SEC61, gamma subunit |
| chr11_+_49434276 | 0.36 |
ENSMUST00000213256.2
|
Olfr1381
|
olfactory receptor 1381 |
| chr9_+_39932760 | 0.35 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr18_-_62313019 | 0.35 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr13_+_109397184 | 0.35 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr11_-_70242850 | 0.33 |
ENSMUST00000019068.7
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr17_+_33483650 | 0.32 |
ENSMUST00000217023.3
|
Olfr63
|
olfactory receptor 63 |
| chr5_+_122781941 | 0.31 |
ENSMUST00000100737.10
ENSMUST00000121489.8 ENSMUST00000031425.15 ENSMUST00000086247.6 |
P2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr13_+_17869988 | 0.30 |
ENSMUST00000049744.4
|
Mplkip
|
M-phase specific PLK1 intereacting protein |
| chr9_-_49397508 | 0.30 |
ENSMUST00000055096.5
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr14_+_63359911 | 0.28 |
ENSMUST00000006235.9
|
Ctsb
|
cathepsin B |
| chr2_+_136733421 | 0.28 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr14_+_58035640 | 0.27 |
ENSMUST00000111269.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr9_-_56325344 | 0.26 |
ENSMUST00000061552.15
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr17_-_10501816 | 0.26 |
ENSMUST00000233684.2
|
Qk
|
quaking, KH domain containing RNA binding |
| chr9_-_37525009 | 0.25 |
ENSMUST00000002013.11
|
Spa17
|
sperm autoantigenic protein 17 |
| chr11_+_62349238 | 0.24 |
ENSMUST00000014389.6
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr5_+_31771281 | 0.24 |
ENSMUST00000031024.14
ENSMUST00000202033.2 |
Mrpl33
Gm43809
|
mitochondrial ribosomal protein L33 predicted gene 43809 |
| chr6_+_135339929 | 0.24 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chrM_+_10167 | 0.24 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr17_-_29226886 | 0.24 |
ENSMUST00000232723.2
|
Stk38
|
serine/threonine kinase 38 |
| chr9_-_88320937 | 0.24 |
ENSMUST00000173011.9
|
Snx14
|
sorting nexin 14 |
| chr4_-_124587340 | 0.24 |
ENSMUST00000030738.8
|
Utp11
|
UTP11 small subunit processome component |
| chr11_-_62348599 | 0.23 |
ENSMUST00000127471.9
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr16_+_48692976 | 0.23 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr10_-_129463803 | 0.23 |
ENSMUST00000204979.3
|
Olfr798
|
olfactory receptor 798 |
| chr2_+_87185159 | 0.22 |
ENSMUST00000215163.3
|
Olfr1120
|
olfactory receptor 1120 |
| chr6_-_149090146 | 0.22 |
ENSMUST00000095319.10
ENSMUST00000141346.2 ENSMUST00000111535.8 |
Amn1
|
antagonist of mitotic exit network 1 |
| chr9_-_88320991 | 0.21 |
ENSMUST00000239462.2
ENSMUST00000165315.9 ENSMUST00000173039.9 |
Snx14
|
sorting nexin 14 |
| chr2_+_71042172 | 0.21 |
ENSMUST00000081710.12
|
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr8_+_26091607 | 0.21 |
ENSMUST00000155861.8
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr9_-_37259690 | 0.21 |
ENSMUST00000213633.2
|
Ccdc15
|
coiled-coil domain containing 15 |
| chr3_+_103767581 | 0.19 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr6_+_86986818 | 0.19 |
ENSMUST00000032060.15
ENSMUST00000117583.8 ENSMUST00000144776.7 |
Nfu1
|
NFU1 iron-sulfur cluster scaffold |
| chrX_+_67722147 | 0.19 |
ENSMUST00000088546.12
|
Fmr1
|
FMRP translational regulator 1 |
| chr11_-_83193412 | 0.19 |
ENSMUST00000176374.2
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr4_+_136013372 | 0.18 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr4_-_132260799 | 0.18 |
ENSMUST00000152993.8
ENSMUST00000067496.7 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr2_+_24076481 | 0.18 |
ENSMUST00000057567.3
|
Il1f9
|
interleukin 1 family, member 9 |
| chr2_+_71042285 | 0.18 |
ENSMUST00000112144.9
ENSMUST00000100028.10 ENSMUST00000112136.2 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr2_-_163913014 | 0.17 |
ENSMUST00000018466.4
ENSMUST00000109384.10 |
Tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr10_+_80084955 | 0.17 |
ENSMUST00000105364.8
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr12_+_84147571 | 0.17 |
ENSMUST00000222921.2
|
Acot6
|
acyl-CoA thioesterase 6 |
| chr5_-_3697806 | 0.16 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr9_-_21504032 | 0.16 |
ENSMUST00000078572.9
|
Yipf2
|
Yip1 domain family, member 2 |
| chr2_+_71042050 | 0.16 |
ENSMUST00000112142.8
ENSMUST00000112139.8 ENSMUST00000112140.8 ENSMUST00000112138.8 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr19_-_40502117 | 0.16 |
ENSMUST00000225148.2
ENSMUST00000224667.2 ENSMUST00000225153.2 ENSMUST00000238831.2 ENSMUST00000226047.2 ENSMUST00000224247.2 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr7_-_132201344 | 0.16 |
ENSMUST00000054562.4
|
Nkx1-2
|
NK1 homeobox 2 |
| chr14_+_73475335 | 0.16 |
ENSMUST00000044405.8
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chrX_-_165992311 | 0.16 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr9_-_88320962 | 0.16 |
ENSMUST00000174806.9
|
Snx14
|
sorting nexin 14 |
| chr12_-_28685913 | 0.15 |
ENSMUST00000074267.5
|
Rps7
|
ribosomal protein S7 |
| chr8_-_11728727 | 0.15 |
ENSMUST00000033906.11
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr5_+_138228074 | 0.15 |
ENSMUST00000159798.8
ENSMUST00000159964.2 |
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr9_-_21504101 | 0.15 |
ENSMUST00000180365.9
ENSMUST00000213809.2 ENSMUST00000034700.15 |
Yipf2
|
Yip1 domain family, member 2 |
| chrX_-_165293419 | 0.15 |
ENSMUST00000112188.8
|
Tceanc
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr7_+_100021425 | 0.14 |
ENSMUST00000098259.11
ENSMUST00000051777.15 |
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr16_+_13572409 | 0.14 |
ENSMUST00000239037.2
|
A930007A09Rik
|
RIKEN cDNA A930007A09 gene |
| chr16_+_65317389 | 0.14 |
ENSMUST00000176330.8
ENSMUST00000004964.15 ENSMUST00000176038.8 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr10_-_33500583 | 0.14 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr7_-_80338600 | 0.14 |
ENSMUST00000122255.8
|
Crtc3
|
CREB regulated transcription coactivator 3 |
| chr9_-_19452372 | 0.14 |
ENSMUST00000213834.2
|
Olfr853
|
olfactory receptor 853 |
| chrX_-_20955370 | 0.13 |
ENSMUST00000040667.13
|
Zfp300
|
zinc finger protein 300 |
| chr6_+_57180275 | 0.13 |
ENSMUST00000226892.2
ENSMUST00000227421.2 |
Vmn1r13
|
vomeronasal 1 receptor 13 |
| chr11_-_53321606 | 0.13 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr16_-_58898368 | 0.13 |
ENSMUST00000216495.3
|
Olfr190
|
olfactory receptor 190 |
| chr2_+_109848224 | 0.13 |
ENSMUST00000150183.9
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr7_-_24423715 | 0.13 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr16_-_36695166 | 0.12 |
ENSMUST00000075946.12
|
Eaf2
|
ELL associated factor 2 |
| chr9_+_120400510 | 0.12 |
ENSMUST00000165532.3
|
Rpl14
|
ribosomal protein L14 |
| chr2_+_89757653 | 0.12 |
ENSMUST00000213720.3
ENSMUST00000102609.3 |
Olfr1258
|
olfactory receptor 1258 |
| chr13_+_41013230 | 0.12 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr7_-_29605949 | 0.12 |
ENSMUST00000159920.2
ENSMUST00000162592.8 |
Zfp27
|
zinc finger protein 27 |
| chr14_+_4198620 | 0.12 |
ENSMUST00000080281.14
|
Rpl15
|
ribosomal protein L15 |
| chr7_+_126376099 | 0.11 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr5_+_150597204 | 0.11 |
ENSMUST00000202170.4
ENSMUST00000016569.11 |
Pds5b
|
PDS5 cohesin associated factor B |
| chr14_+_58035614 | 0.11 |
ENSMUST00000128764.9
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr15_-_59245998 | 0.11 |
ENSMUST00000022976.6
|
Washc5
|
WASH complex subunit 5 |
| chr4_+_141148068 | 0.11 |
ENSMUST00000102486.5
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr8_-_31658775 | 0.11 |
ENSMUST00000033983.6
|
Mak16
|
MAK16 homolog |
| chr4_-_129334593 | 0.11 |
ENSMUST00000053042.6
ENSMUST00000106046.8 |
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr1_+_82702598 | 0.11 |
ENSMUST00000078332.13
ENSMUST00000073025.12 ENSMUST00000161648.8 ENSMUST00000160786.8 ENSMUST00000162003.8 |
Mff
|
mitochondrial fission factor |
| chr10_+_80085275 | 0.11 |
ENSMUST00000020361.7
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr5_+_137015873 | 0.11 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr11_+_115921129 | 0.10 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr7_-_43906802 | 0.10 |
ENSMUST00000107945.8
ENSMUST00000118216.8 |
Acp4
|
acid phosphatase 4 |
| chr4_+_63133639 | 0.10 |
ENSMUST00000036300.13
|
Col27a1
|
collagen, type XXVII, alpha 1 |
| chr3_+_122068018 | 0.10 |
ENSMUST00000035776.10
|
Dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr17_-_47922374 | 0.10 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr12_-_69403892 | 0.10 |
ENSMUST00000222065.2
ENSMUST00000021368.10 |
Nemf
|
nuclear export mediator factor |
| chr6_+_90078412 | 0.10 |
ENSMUST00000089417.8
ENSMUST00000226577.2 |
Vmn1r50
|
vomeronasal 1 receptor 50 |
| chr14_-_119336809 | 0.10 |
ENSMUST00000156203.8
|
Uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr13_-_67081138 | 0.10 |
ENSMUST00000021991.11
|
Mterf3
|
mitochondrial transcription termination factor 3 |
| chr4_+_40948407 | 0.10 |
ENSMUST00000030128.6
|
Chmp5
|
charged multivesicular body protein 5 |
| chr4_+_155709275 | 0.10 |
ENSMUST00000067081.10
ENSMUST00000105600.8 ENSMUST00000105598.2 |
Cdk11b
|
cyclin-dependent kinase 11B |
| chr13_-_35211060 | 0.10 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr15_-_28025920 | 0.10 |
ENSMUST00000090247.7
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr17_-_7050145 | 0.09 |
ENSMUST00000064234.7
|
Ezr
|
ezrin |
| chr9_+_80072274 | 0.09 |
ENSMUST00000035889.15
ENSMUST00000113268.8 |
Myo6
|
myosin VI |
| chrX_-_41000746 | 0.09 |
ENSMUST00000047037.15
|
Thoc2
|
THO complex 2 |
| chrX_-_165992145 | 0.09 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr16_-_14135183 | 0.09 |
ENSMUST00000120707.2
ENSMUST00000023357.14 |
Cep20
|
centrosomal protein 20 |
| chr1_+_155433858 | 0.09 |
ENSMUST00000080138.13
ENSMUST00000035560.9 ENSMUST00000097529.5 |
Acbd6
|
acyl-Coenzyme A binding domain containing 6 |
| chr8_-_87307294 | 0.09 |
ENSMUST00000131423.8
ENSMUST00000152438.2 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr14_-_51162346 | 0.09 |
ENSMUST00000159292.8
|
Osgep
|
O-sialoglycoprotein endopeptidase |
| chr16_+_33201227 | 0.08 |
ENSMUST00000232023.2
|
Zfp148
|
zinc finger protein 148 |
| chr5_+_146885450 | 0.08 |
ENSMUST00000146511.8
ENSMUST00000132102.2 |
Gtf3a
|
general transcription factor III A |
| chr10_+_78864575 | 0.08 |
ENSMUST00000203906.3
|
Olfr57
|
olfactory receptor 57 |
| chr18_+_37580692 | 0.08 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr15_+_38869667 | 0.08 |
ENSMUST00000022906.8
|
Fzd6
|
frizzled class receptor 6 |
| chr15_+_98367777 | 0.08 |
ENSMUST00000216180.2
|
OR5BS1P
|
olfactory receptor 240, pseudogene 1 |
| chr4_-_135749032 | 0.08 |
ENSMUST00000030427.6
|
Eloa
|
elongin A |
| chr6_+_34757771 | 0.07 |
ENSMUST00000115012.8
ENSMUST00000115014.8 ENSMUST00000115009.2 |
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr8_-_106198112 | 0.07 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr16_+_33201164 | 0.07 |
ENSMUST00000165418.9
|
Zfp148
|
zinc finger protein 148 |
| chr4_-_129512862 | 0.07 |
ENSMUST00000121442.2
ENSMUST00000046675.12 |
Iqcc
|
IQ motif containing C |
| chr11_-_102298748 | 0.07 |
ENSMUST00000124755.2
|
Slc25a39
|
solute carrier family 25, member 39 |
| chr17_-_29226700 | 0.07 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38 |
| chr4_-_14826587 | 0.06 |
ENSMUST00000117268.9
ENSMUST00000236953.2 |
Otud6b
|
OTU domain containing 6B |
| chrM_+_7779 | 0.06 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr8_+_118225008 | 0.06 |
ENSMUST00000081232.9
|
Plcg2
|
phospholipase C, gamma 2 |
| chr15_+_39997761 | 0.06 |
ENSMUST00000228780.2
|
9330182O14Rik
|
RIKEN cDNA 9330182O14 gene |
| chr6_-_66620810 | 0.06 |
ENSMUST00000228647.2
ENSMUST00000227332.2 ENSMUST00000226262.2 ENSMUST00000226999.2 |
Vmn1r34
|
vomeronasal 1 receptor 34 |
| chr3_-_116456292 | 0.06 |
ENSMUST00000029570.9
|
Mfsd14a
|
major facilitator superfamily domain containing 14A |
| chr12_-_110649040 | 0.06 |
ENSMUST00000222915.2
ENSMUST00000070659.7 |
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr3_+_90155479 | 0.06 |
ENSMUST00000015467.9
|
Slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr2_+_107120934 | 0.06 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr13_+_117356808 | 0.06 |
ENSMUST00000022242.9
|
Emb
|
embigin |
| chr8_-_26087552 | 0.06 |
ENSMUST00000210234.2
ENSMUST00000211422.2 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr8_-_71349927 | 0.06 |
ENSMUST00000212709.2
ENSMUST00000212796.2 ENSMUST00000212378.2 ENSMUST00000054220.10 ENSMUST00000212494.2 |
Rpl18a
|
ribosomal protein L18A |
| chr4_+_129714494 | 0.06 |
ENSMUST00000165853.2
|
Ptp4a2
|
protein tyrosine phosphatase 4a2 |
| chr13_+_49806772 | 0.05 |
ENSMUST00000223264.2
ENSMUST00000221142.2 ENSMUST00000222333.2 ENSMUST00000021824.8 |
Nol8
|
nucleolar protein 8 |
| chr14_-_52733146 | 0.05 |
ENSMUST00000206931.2
ENSMUST00000206062.4 |
Olfr1507
|
olfactory receptor 1507 |
| chr11_+_97206542 | 0.05 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr17_-_23086038 | 0.05 |
ENSMUST00000088696.13
|
Zfp945
|
zinc finger protein 945 |
| chr15_-_4025190 | 0.05 |
ENSMUST00000046633.10
|
AW549877
|
expressed sequence AW549877 |
| chr13_+_92562404 | 0.05 |
ENSMUST00000061594.13
|
Ankrd34b
|
ankyrin repeat domain 34B |
| chr14_-_119160497 | 0.05 |
ENSMUST00000047208.12
|
Dzip1
|
DAZ interacting protein 1 |
| chrX_+_84913911 | 0.05 |
ENSMUST00000113976.2
|
Tasl
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr12_+_85871691 | 0.05 |
ENSMUST00000040273.14
ENSMUST00000110224.8 ENSMUST00000142411.9 |
Ttll5
|
tubulin tyrosine ligase-like family, member 5 |
| chr5_+_150597292 | 0.05 |
ENSMUST00000038900.15
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr13_+_49806542 | 0.05 |
ENSMUST00000222197.2
ENSMUST00000221083.2 ENSMUST00000223467.2 |
Nol8
|
nucleolar protein 8 |
| chr10_-_81037878 | 0.05 |
ENSMUST00000005069.8
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr14_+_58036093 | 0.05 |
ENSMUST00000111267.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr7_+_43256593 | 0.04 |
ENSMUST00000120935.2
ENSMUST00000127765.8 ENSMUST00000032661.14 |
Zfp819
|
zinc finger protein 819 |
| chr13_-_58506890 | 0.04 |
ENSMUST00000225388.2
|
Kif27
|
kinesin family member 27 |
| chr2_-_118380149 | 0.04 |
ENSMUST00000090219.13
|
Bmf
|
BCL2 modifying factor |
| chr7_+_40636967 | 0.04 |
ENSMUST00000206529.2
ENSMUST00000171664.2 |
4930433I11Rik
|
RIKEN cDNA 4930433I11 gene |
| chr15_-_64254754 | 0.04 |
ENSMUST00000177374.8
ENSMUST00000110114.10 ENSMUST00000110115.9 ENSMUST00000023008.16 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr11_+_78067537 | 0.04 |
ENSMUST00000098545.12
|
Tlcd1
|
TLC domain containing 1 |
| chr2_-_168609110 | 0.04 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr13_-_104365310 | 0.04 |
ENSMUST00000022226.6
|
Ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr17_-_23086085 | 0.04 |
ENSMUST00000160457.10
|
Zfp945
|
zinc finger protein 945 |
| chr17_-_11059172 | 0.04 |
ENSMUST00000041463.7
|
Pacrg
|
PARK2 co-regulated |
| chr13_-_67648563 | 0.04 |
ENSMUST00000163534.2
ENSMUST00000091523.9 ENSMUST00000171518.8 ENSMUST00000076123.12 |
Zfp58
|
zinc finger protein 58 |
| chr7_-_29605504 | 0.04 |
ENSMUST00000053521.15
|
Zfp27
|
zinc finger protein 27 |
| chr6_+_96090127 | 0.04 |
ENSMUST00000122120.8
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr9_+_3403592 | 0.04 |
ENSMUST00000027027.7
|
Cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr3_+_62245765 | 0.04 |
ENSMUST00000079300.13
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chrX_+_106711562 | 0.04 |
ENSMUST00000168174.9
|
Tbx22
|
T-box 22 |
| chr7_+_102731092 | 0.03 |
ENSMUST00000214215.2
|
Olfr584
|
olfactory receptor 584 |
| chr12_+_98737274 | 0.03 |
ENSMUST00000021399.9
|
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr15_-_85695855 | 0.03 |
ENSMUST00000134631.8
ENSMUST00000154814.2 ENSMUST00000071876.13 ENSMUST00000150995.8 |
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr17_+_11059248 | 0.03 |
ENSMUST00000191124.7
|
Prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr4_-_126096551 | 0.03 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr6_-_118174679 | 0.03 |
ENSMUST00000088790.9
|
Ret
|
ret proto-oncogene |
| chr9_+_70450024 | 0.03 |
ENSMUST00000216816.2
|
Sltm
|
SAFB-like, transcription modulator |
| chr11_-_78642480 | 0.03 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr5_+_63806451 | 0.03 |
ENSMUST00000159584.3
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr5_+_9163244 | 0.03 |
ENSMUST00000198935.2
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr11_+_5519677 | 0.03 |
ENSMUST00000109856.8
ENSMUST00000109855.8 ENSMUST00000118112.9 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr15_+_100178718 | 0.03 |
ENSMUST00000096200.6
|
Tmprss12
|
transmembrane (C-terminal) protease, serine 12 |
| chr15_-_98832403 | 0.03 |
ENSMUST00000077577.8
|
Tuba1b
|
tubulin, alpha 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sp100
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:2000387 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.3 | 0.9 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.3 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.1 | 0.3 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.1 | 0.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 1.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.2 | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 0.2 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.2 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.5 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.0 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:1904667 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0019034 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.0 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 1.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.4 | GO:0031711 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.3 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 1.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.9 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |