Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Sp2
Z-value: 0.18
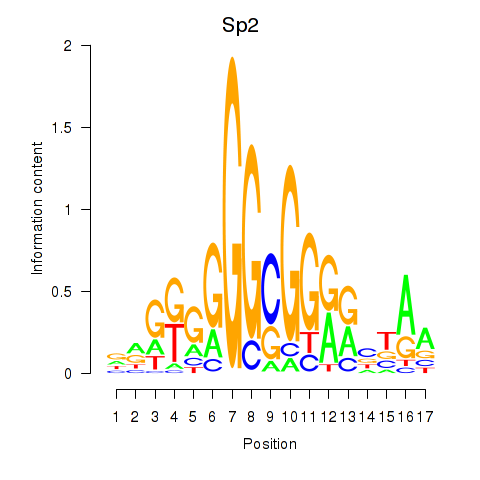
Transcription factors associated with Sp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp2
|
ENSMUSG00000018678.14 | Sp2 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp2 | mm39_v1_chr11_-_96868483_96868537 | -0.38 | 5.2e-01 | Click! |
Activity profile of Sp2 motif
Sorted Z-values of Sp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_14411956 | 0.12 |
ENSMUST00000215143.2
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr10_+_80003612 | 0.11 |
ENSMUST00000105365.9
|
Cirbp
|
cold inducible RNA binding protein |
| chr3_+_89987749 | 0.11 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chr17_+_34135145 | 0.10 |
ENSMUST00000053429.11
ENSMUST00000234582.2 |
Zbtb22
Tapbp
|
zinc finger and BTB domain containing 22 TAP binding protein |
| chr14_+_55815879 | 0.09 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr12_+_112586501 | 0.09 |
ENSMUST00000180015.9
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr18_+_60907698 | 0.09 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr11_-_70537690 | 0.09 |
ENSMUST00000136383.2
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr12_-_36206780 | 0.08 |
ENSMUST00000223382.2
ENSMUST00000020856.6 |
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr12_-_36206626 | 0.08 |
ENSMUST00000220828.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr7_-_26878260 | 0.08 |
ENSMUST00000093040.13
|
Rab4b
|
RAB4B, member RAS oncogene family |
| chr18_+_60907668 | 0.08 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr18_-_35795233 | 0.07 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr14_-_55828511 | 0.07 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr13_-_111626562 | 0.06 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr6_+_51447613 | 0.06 |
ENSMUST00000114445.8
ENSMUST00000114446.8 ENSMUST00000141711.3 |
Cbx3
|
chromobox 3 |
| chr12_+_112586465 | 0.06 |
ENSMUST00000021726.8
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr14_+_118374511 | 0.06 |
ENSMUST00000022728.4
|
Gpr180
|
G protein-coupled receptor 180 |
| chr3_+_146205864 | 0.06 |
ENSMUST00000119130.2
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr10_+_128173603 | 0.06 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr18_-_38088062 | 0.06 |
ENSMUST00000043498.9
|
Hdac3
|
histone deacetylase 3 |
| chr3_+_94882142 | 0.06 |
ENSMUST00000167008.8
ENSMUST00000107251.9 |
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr1_-_65225617 | 0.06 |
ENSMUST00000186222.7
ENSMUST00000169032.8 ENSMUST00000191459.2 ENSMUST00000188876.7 |
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr14_+_55815817 | 0.06 |
ENSMUST00000174259.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr11_+_97689819 | 0.05 |
ENSMUST00000143571.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr3_-_108053396 | 0.05 |
ENSMUST00000000001.5
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr11_-_69872050 | 0.05 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chrX_-_72502595 | 0.05 |
ENSMUST00000033737.15
ENSMUST00000077243.5 |
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr3_-_101511971 | 0.05 |
ENSMUST00000036493.8
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr19_+_4560500 | 0.05 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr7_-_29931612 | 0.05 |
ENSMUST00000006254.6
|
Tbcb
|
tubulin folding cofactor B |
| chr19_-_4977997 | 0.05 |
ENSMUST00000236758.2
|
Dpp3
|
dipeptidylpeptidase 3 |
| chr7_-_4687916 | 0.05 |
ENSMUST00000206306.2
ENSMUST00000205952.2 ENSMUST00000079970.6 |
Hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr5_-_137529251 | 0.05 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr7_+_45367545 | 0.05 |
ENSMUST00000209287.2
|
Rpl18
|
ribosomal protein L18 |
| chr12_-_36206750 | 0.05 |
ENSMUST00000221388.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr5_+_136982100 | 0.05 |
ENSMUST00000111094.8
ENSMUST00000111097.8 |
Fis1
|
fission, mitochondrial 1 |
| chr4_+_3938881 | 0.05 |
ENSMUST00000108386.8
ENSMUST00000121110.8 ENSMUST00000149544.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr8_+_85696453 | 0.05 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr9_+_83807162 | 0.05 |
ENSMUST00000190637.7
ENSMUST00000034801.11 |
Bckdhb
|
branched chain ketoacid dehydrogenase E1, beta polypeptide |
| chrX_-_168103266 | 0.05 |
ENSMUST00000033717.9
ENSMUST00000112115.2 |
Hccs
|
holocytochrome c synthetase |
| chr17_-_56447332 | 0.05 |
ENSMUST00000001256.11
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr7_+_142606476 | 0.04 |
ENSMUST00000037941.10
|
Cd81
|
CD81 antigen |
| chr7_+_127345909 | 0.04 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr11_-_73067828 | 0.04 |
ENSMUST00000108480.2
ENSMUST00000054952.4 |
Emc6
|
ER membrane protein complex subunit 6 |
| chr15_-_63869818 | 0.04 |
ENSMUST00000164532.3
|
Cyrib
|
CYFIP related Rac1 interactor B |
| chr11_+_80191692 | 0.04 |
ENSMUST00000017836.8
|
Rhbdl3
|
rhomboid like 3 |
| chr4_+_63462984 | 0.04 |
ENSMUST00000035301.7
|
Atp6v1g1
|
ATPase, H+ transporting, lysosomal V1 subunit G1 |
| chr2_+_131028861 | 0.04 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr14_-_31552335 | 0.04 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr6_-_119825081 | 0.04 |
ENSMUST00000183703.8
ENSMUST00000183911.8 |
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr14_-_123220554 | 0.04 |
ENSMUST00000126867.8
ENSMUST00000148661.2 ENSMUST00000037726.14 |
Tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr1_-_65225572 | 0.04 |
ENSMUST00000188109.7
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr6_-_30873669 | 0.04 |
ENSMUST00000048774.13
ENSMUST00000166192.7 |
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chrX_+_12454031 | 0.04 |
ENSMUST00000033313.3
|
Atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr4_+_148215339 | 0.04 |
ENSMUST00000084129.9
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr9_-_64160899 | 0.04 |
ENSMUST00000005066.9
|
Map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr19_-_8775817 | 0.04 |
ENSMUST00000235964.2
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr9_-_21202545 | 0.04 |
ENSMUST00000215619.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr2_-_37249247 | 0.04 |
ENSMUST00000112940.8
ENSMUST00000009174.15 |
Pdcl
|
phosducin-like |
| chr4_-_133273243 | 0.04 |
ENSMUST00000030665.7
|
Nudc
|
nudC nuclear distribution protein |
| chr7_+_141027763 | 0.04 |
ENSMUST00000106003.2
|
Rplp2
|
ribosomal protein, large P2 |
| chr15_+_78312764 | 0.04 |
ENSMUST00000162517.8
ENSMUST00000166142.10 ENSMUST00000089414.11 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr8_-_73324877 | 0.04 |
ENSMUST00000058733.9
ENSMUST00000167290.8 |
Smim7
|
small integral membrane protein 7 |
| chr7_+_19242634 | 0.04 |
ENSMUST00000002112.15
ENSMUST00000108455.8 |
Trappc6a
|
trafficking protein particle complex 6A |
| chr3_+_138148846 | 0.04 |
ENSMUST00000005964.7
|
Adh5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr2_+_163836880 | 0.04 |
ENSMUST00000018470.10
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
| chr13_-_58276353 | 0.04 |
ENSMUST00000007980.7
|
Hnrnpa0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr19_+_44994094 | 0.04 |
ENSMUST00000236685.2
|
Twnk
|
twinkle mtDNA helicase |
| chr15_+_88746380 | 0.04 |
ENSMUST00000042818.11
|
Pim3
|
proviral integration site 3 |
| chr3_+_94882038 | 0.04 |
ENSMUST00000072287.12
|
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr10_-_112764879 | 0.04 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr7_-_26866157 | 0.04 |
ENSMUST00000080058.11
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr18_-_66155651 | 0.04 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr11_-_58059293 | 0.04 |
ENSMUST00000172035.8
ENSMUST00000035604.13 ENSMUST00000102711.9 |
Gemin5
|
gem nuclear organelle associated protein 5 |
| chr19_+_8944369 | 0.04 |
ENSMUST00000052248.8
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr5_-_140816111 | 0.04 |
ENSMUST00000000153.9
|
Gna12
|
guanine nucleotide binding protein, alpha 12 |
| chr13_-_95511837 | 0.04 |
ENSMUST00000022189.9
|
Aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr11_-_68864666 | 0.04 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr1_-_16727242 | 0.04 |
ENSMUST00000186948.7
ENSMUST00000187910.7 ENSMUST00000115352.10 |
Eloc
|
elongin C |
| chr11_-_113641980 | 0.04 |
ENSMUST00000153453.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr7_+_27186335 | 0.04 |
ENSMUST00000008528.8
|
Sertad1
|
SERTA domain containing 1 |
| chr4_+_28813125 | 0.04 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr4_-_87951565 | 0.04 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr9_+_109704609 | 0.04 |
ENSMUST00000094324.8
|
Cdc25a
|
cell division cycle 25A |
| chr9_+_108673171 | 0.03 |
ENSMUST00000195514.6
ENSMUST00000085018.6 ENSMUST00000192028.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr2_+_163837423 | 0.03 |
ENSMUST00000131288.2
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
| chr12_+_103498542 | 0.03 |
ENSMUST00000021631.12
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr9_-_21202693 | 0.03 |
ENSMUST00000213407.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr15_-_79658584 | 0.03 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr2_+_181139016 | 0.03 |
ENSMUST00000108799.10
|
Tpd52l2
|
tumor protein D52-like 2 |
| chr8_-_84831391 | 0.03 |
ENSMUST00000041367.9
ENSMUST00000210279.2 |
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr14_-_31552608 | 0.03 |
ENSMUST00000014640.9
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr4_+_107736942 | 0.03 |
ENSMUST00000030348.6
|
Magoh
|
mago homolog, exon junction complex core component |
| chrX_+_141011173 | 0.03 |
ENSMUST00000112914.8
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr2_+_125708027 | 0.03 |
ENSMUST00000134337.8
ENSMUST00000094604.9 |
Galk2
|
galactokinase 2 |
| chr2_+_71219561 | 0.03 |
ENSMUST00000028408.3
|
Hat1
|
histone aminotransferase 1 |
| chr18_-_35795175 | 0.03 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr6_-_124391994 | 0.03 |
ENSMUST00000035861.6
ENSMUST00000112532.8 ENSMUST00000080557.12 |
Pex5
|
peroxisomal biogenesis factor 5 |
| chr16_-_91525655 | 0.03 |
ENSMUST00000117644.8
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr19_-_4978238 | 0.03 |
ENSMUST00000237394.2
ENSMUST00000025851.4 |
Dpp3
|
dipeptidylpeptidase 3 |
| chr8_+_12965876 | 0.03 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr14_+_21102662 | 0.03 |
ENSMUST00000223915.2
|
Adk
|
adenosine kinase |
| chr14_-_31362835 | 0.03 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr7_-_44711075 | 0.03 |
ENSMUST00000007981.9
ENSMUST00000210500.2 ENSMUST00000210493.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr1_-_160620478 | 0.03 |
ENSMUST00000195442.6
ENSMUST00000028049.13 |
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr16_-_91525863 | 0.03 |
ENSMUST00000073466.13
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr4_+_41760454 | 0.03 |
ENSMUST00000108040.8
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr10_+_75353844 | 0.03 |
ENSMUST00000220229.2
|
Snrpd3
|
small nuclear ribonucleoprotein D3 |
| chr4_-_108436514 | 0.03 |
ENSMUST00000079213.6
|
Prpf38a
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing A |
| chr19_-_5474509 | 0.03 |
ENSMUST00000136579.3
ENSMUST00000113673.9 |
Drap1
|
Dr1 associated protein 1 (negative cofactor 2 alpha) |
| chr8_-_85696040 | 0.03 |
ENSMUST00000214133.2
ENSMUST00000147812.8 |
Gm49661
Rnaseh2a
|
predicted gene, 49661 ribonuclease H2, large subunit |
| chr7_+_142558837 | 0.03 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chr4_-_155976158 | 0.03 |
ENSMUST00000097737.5
|
Pusl1
|
pseudouridylate synthase-like 1 |
| chr4_-_16013867 | 0.03 |
ENSMUST00000037198.10
|
Osgin2
|
oxidative stress induced growth inhibitor family member 2 |
| chr15_-_79658608 | 0.03 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr9_-_21202353 | 0.03 |
ENSMUST00000086374.8
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr11_+_87938128 | 0.03 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr3_+_107803563 | 0.03 |
ENSMUST00000169365.2
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr18_+_38088597 | 0.03 |
ENSMUST00000070709.9
ENSMUST00000177058.8 ENSMUST00000169360.9 ENSMUST00000163591.9 ENSMUST00000091932.12 |
Rell2
|
RELT-like 2 |
| chr19_+_44994905 | 0.03 |
ENSMUST00000026227.3
|
Twnk
|
twinkle mtDNA helicase |
| chrX_-_162859429 | 0.03 |
ENSMUST00000134272.2
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B |
| chr7_+_45367479 | 0.03 |
ENSMUST00000072503.13
ENSMUST00000211061.2 ENSMUST00000209693.2 |
Rpl18
|
ribosomal protein L18 |
| chr4_+_138032035 | 0.03 |
ENSMUST00000030538.5
|
Ddost
|
dolichyl-di-phosphooligosaccharide-protein glycotransferase |
| chr12_-_21467437 | 0.03 |
ENSMUST00000103002.8
ENSMUST00000155480.9 ENSMUST00000135088.9 |
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| chr12_-_102709884 | 0.03 |
ENSMUST00000173760.9
ENSMUST00000178384.2 |
Moap1
|
modulator of apoptosis 1 |
| chr11_-_70860778 | 0.03 |
ENSMUST00000108530.2
ENSMUST00000035283.11 ENSMUST00000108531.8 |
Nup88
|
nucleoporin 88 |
| chr19_-_6957789 | 0.03 |
ENSMUST00000070878.9
ENSMUST00000177752.9 |
Fkbp2
|
FK506 binding protein 2 |
| chr10_+_127871493 | 0.03 |
ENSMUST00000092048.13
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr19_+_6107874 | 0.03 |
ENSMUST00000178310.9
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr8_+_85696396 | 0.03 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr13_-_58421935 | 0.03 |
ENSMUST00000091579.6
|
Gkap1
|
G kinase anchoring protein 1 |
| chr2_+_30697838 | 0.03 |
ENSMUST00000041830.10
ENSMUST00000152374.8 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr2_+_30872291 | 0.03 |
ENSMUST00000102849.11
|
Usp20
|
ubiquitin specific peptidase 20 |
| chr10_+_127126643 | 0.03 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr1_-_183766195 | 0.03 |
ENSMUST00000050306.8
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chrX_+_141010919 | 0.03 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr13_-_38842967 | 0.03 |
ENSMUST00000001757.9
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr9_+_108216233 | 0.03 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr11_+_87938519 | 0.03 |
ENSMUST00000079866.11
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr17_-_34174631 | 0.03 |
ENSMUST00000174609.9
ENSMUST00000008812.9 |
Rps18
|
ribosomal protein S18 |
| chr14_+_21102642 | 0.03 |
ENSMUST00000045376.11
|
Adk
|
adenosine kinase |
| chr12_+_3941728 | 0.03 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr14_+_119175328 | 0.03 |
ENSMUST00000022734.9
|
Dnajc3
|
DnaJ heat shock protein family (Hsp40) member C3 |
| chr19_-_44994824 | 0.03 |
ENSMUST00000097715.4
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr7_-_44711130 | 0.03 |
ENSMUST00000211337.2
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr15_-_79718423 | 0.03 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chrX_+_158155171 | 0.03 |
ENSMUST00000087143.7
|
Eif1ax
|
eukaryotic translation initiation factor 1A, X-linked |
| chr4_-_114844417 | 0.03 |
ENSMUST00000030491.9
|
Cmpk1
|
cytidine monophosphate (UMP-CMP) kinase 1 |
| chr12_+_112611322 | 0.02 |
ENSMUST00000109755.5
|
Siva1
|
SIVA1, apoptosis-inducing factor |
| chr12_+_32428691 | 0.02 |
ENSMUST00000172332.4
|
Ccdc71l
|
coiled-coil domain containing 71 like |
| chr9_-_103243039 | 0.02 |
ENSMUST00000035484.11
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr8_-_110419867 | 0.02 |
ENSMUST00000034164.6
|
Ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr15_+_76580925 | 0.02 |
ENSMUST00000023203.6
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr6_+_7844759 | 0.02 |
ENSMUST00000040159.6
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr7_+_45084300 | 0.02 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr7_+_101545547 | 0.02 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr7_-_26865838 | 0.02 |
ENSMUST00000108382.2
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr3_-_8988854 | 0.02 |
ENSMUST00000042148.6
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr11_-_74816484 | 0.02 |
ENSMUST00000138612.2
ENSMUST00000123855.8 ENSMUST00000128556.8 ENSMUST00000108448.8 ENSMUST00000108447.8 ENSMUST00000065211.9 |
Srr
|
serine racemase |
| chr18_-_46730547 | 0.02 |
ENSMUST00000151189.2
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr11_-_95956176 | 0.02 |
ENSMUST00000100528.5
|
Ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr17_+_35827997 | 0.02 |
ENSMUST00000164242.9
ENSMUST00000045956.14 |
Cchcr1
|
coiled-coil alpha-helical rod protein 1 |
| chr3_+_107803225 | 0.02 |
ENSMUST00000172247.8
ENSMUST00000167387.8 |
Gstm5
|
glutathione S-transferase, mu 5 |
| chr17_+_35201074 | 0.02 |
ENSMUST00000007266.14
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr9_+_14411921 | 0.02 |
ENSMUST00000213913.2
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr5_+_100666278 | 0.02 |
ENSMUST00000151414.8
|
Cops4
|
COP9 signalosome subunit 4 |
| chr6_-_119825020 | 0.02 |
ENSMUST00000184864.8
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr19_-_12478803 | 0.02 |
ENSMUST00000045521.9
|
Dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr3_-_89867869 | 0.02 |
ENSMUST00000069805.14
|
Atp8b2
|
ATPase, class I, type 8B, member 2 |
| chr2_-_29938841 | 0.02 |
ENSMUST00000113711.3
|
Dync2i2
|
dynein 2 intermediate chain 2 |
| chr2_-_35939377 | 0.02 |
ENSMUST00000070112.6
|
Ndufa8
|
NADH:ubiquinone oxidoreductase subunit A8 |
| chr15_+_79113341 | 0.02 |
ENSMUST00000163571.8
|
Pick1
|
protein interacting with C kinase 1 |
| chr1_+_93406809 | 0.02 |
ENSMUST00000112912.8
|
Septin2
|
septin 2 |
| chr5_-_137529465 | 0.02 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr6_-_143045493 | 0.02 |
ENSMUST00000204655.3
ENSMUST00000111758.9 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr18_+_35731735 | 0.02 |
ENSMUST00000236868.2
|
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr11_+_105927698 | 0.02 |
ENSMUST00000058438.9
|
Dcaf7
|
DDB1 and CUL4 associated factor 7 |
| chr9_+_65368207 | 0.02 |
ENSMUST00000034955.8
ENSMUST00000213957.2 |
Spg21
|
SPG21, maspardin |
| chr8_+_85598734 | 0.02 |
ENSMUST00000170296.2
ENSMUST00000136026.8 |
Syce2
|
synaptonemal complex central element protein 2 |
| chr19_-_5474934 | 0.02 |
ENSMUST00000113674.8
|
Drap1
|
Dr1 associated protein 1 (negative cofactor 2 alpha) |
| chr16_-_10360893 | 0.02 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr18_+_76374959 | 0.02 |
ENSMUST00000171256.8
|
Smad2
|
SMAD family member 2 |
| chr3_+_68912043 | 0.02 |
ENSMUST00000042901.15
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr16_-_10603389 | 0.02 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr4_-_56865134 | 0.02 |
ENSMUST00000107612.3
ENSMUST00000045142.15 |
Ctnnal1
|
catenin (cadherin associated protein), alpha-like 1 |
| chr1_-_65218217 | 0.02 |
ENSMUST00000097709.11
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr10_-_128401773 | 0.02 |
ENSMUST00000026425.13
ENSMUST00000131728.4 |
Pa2g4
|
proliferation-associated 2G4 |
| chr19_+_6107957 | 0.02 |
ENSMUST00000237859.2
ENSMUST00000043074.14 ENSMUST00000236336.2 ENSMUST00000179142.2 ENSMUST00000236217.2 |
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr8_+_85696695 | 0.02 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr9_-_57552392 | 0.02 |
ENSMUST00000216934.2
|
Csk
|
c-src tyrosine kinase |
| chr15_-_25413838 | 0.02 |
ENSMUST00000058845.9
|
Basp1
|
brain abundant, membrane attached signal protein 1 |
| chr9_-_63306497 | 0.02 |
ENSMUST00000168665.3
|
2300009A05Rik
|
RIKEN cDNA 2300009A05 gene |
| chr9_+_40103598 | 0.02 |
ENSMUST00000026693.14
ENSMUST00000168832.2 |
Zfp202
|
zinc finger protein 202 |
| chr11_-_21522193 | 0.02 |
ENSMUST00000102874.11
ENSMUST00000238916.2 |
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr19_-_5474787 | 0.02 |
ENSMUST00000148219.9
|
Drap1
|
Dr1 associated protein 1 (negative cofactor 2 alpha) |
| chr19_+_42078859 | 0.02 |
ENSMUST00000235932.2
ENSMUST00000066778.6 |
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr17_+_56611313 | 0.02 |
ENSMUST00000113035.8
ENSMUST00000113039.9 ENSMUST00000142387.2 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr11_-_21521934 | 0.02 |
ENSMUST00000239073.2
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chrX_+_8758614 | 0.02 |
ENSMUST00000064196.5
|
B630019K06Rik
|
RIKEN cDNA B630019K06 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0050823 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 0.1 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.0 | GO:0006069 | ethanol oxidation(GO:0006069) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.0 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.0 | GO:0030622 | U4atac snRNA binding(GO:0030622) |