Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
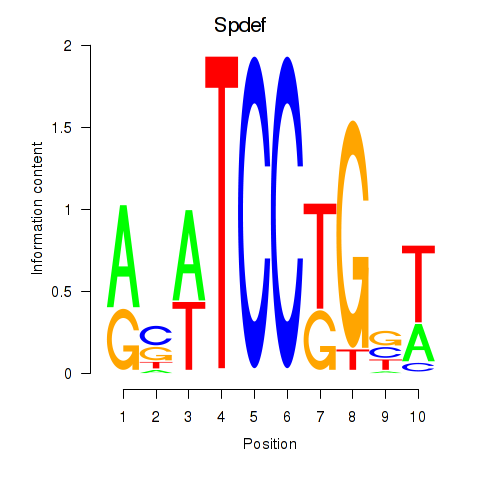
Results for Spdef
Z-value: 2.24
Transcription factors associated with Spdef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spdef
|
ENSMUSG00000024215.15 | SAM pointed domain containing ets transcription factor |
Activity profile of Spdef motif
Sorted Z-values of Spdef motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_22017906 | 2.09 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24 |
| chr13_-_21900313 | 2.06 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr13_+_21906214 | 1.86 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr13_+_22227359 | 1.85 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr2_+_129854256 | 1.75 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr19_+_8975249 | 1.72 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr9_+_57818243 | 1.58 |
ENSMUST00000216925.2
ENSMUST00000163329.2 ENSMUST00000213654.2 ENSMUST00000217132.2 ENSMUST00000216841.2 ENSMUST00000214086.2 |
Ubl7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chr11_+_70538083 | 1.40 |
ENSMUST00000037534.8
|
Rnf167
|
ring finger protein 167 |
| chr14_-_55909314 | 1.37 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr10_+_17598961 | 1.34 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr17_+_34808772 | 1.28 |
ENSMUST00000038244.15
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr6_-_97408367 | 1.17 |
ENSMUST00000124050.3
|
Frmd4b
|
FERM domain containing 4B |
| chr14_-_55909527 | 1.16 |
ENSMUST00000010520.10
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr9_+_20555629 | 1.12 |
ENSMUST00000161887.8
|
Ubl5
|
ubiquitin-like 5 |
| chr16_-_4698148 | 1.05 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr12_-_31549538 | 0.97 |
ENSMUST00000064240.14
ENSMUST00000185739.8 ENSMUST00000188326.3 ENSMUST00000101499.10 ENSMUST00000085487.12 |
Cbll1
|
Casitas B-lineage lymphoma-like 1 |
| chr9_-_43027809 | 0.93 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr13_-_22227114 | 0.91 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr6_+_35229589 | 0.90 |
ENSMUST00000152147.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr8_-_123768984 | 0.88 |
ENSMUST00000212937.2
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr9_-_41068771 | 0.85 |
ENSMUST00000136530.8
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B |
| chr3_-_122778052 | 0.83 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr2_-_168432111 | 0.82 |
ENSMUST00000029057.13
ENSMUST00000074618.10 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr8_-_70963202 | 0.81 |
ENSMUST00000125184.8
|
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr11_-_119246310 | 0.80 |
ENSMUST00000100172.3
ENSMUST00000005173.11 |
Sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr2_-_127050161 | 0.80 |
ENSMUST00000056146.2
|
1810024B03Rik
|
RIKEN cDNA 1810024B03 gene |
| chr14_-_54491365 | 0.75 |
ENSMUST00000128231.2
|
Dad1
|
defender against cell death 1 |
| chr8_+_114362181 | 0.75 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr7_+_19699291 | 0.74 |
ENSMUST00000094753.6
|
Ceacam20
|
carcinoembryonic antigen-related cell adhesion molecule 20 |
| chr8_+_114362419 | 0.73 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr7_-_46569617 | 0.72 |
ENSMUST00000210664.2
ENSMUST00000156335.9 |
Tsg101
|
tumor susceptibility gene 101 |
| chr16_-_92622972 | 0.70 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr7_+_127078371 | 0.70 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chr5_-_31065036 | 0.70 |
ENSMUST00000132034.5
ENSMUST00000132253.5 |
Ost4
|
oligosaccharyltransferase complex subunit 4 (non-catalytic) |
| chr11_-_86574586 | 0.70 |
ENSMUST00000018315.10
|
Vmp1
|
vacuole membrane protein 1 |
| chr7_+_27878894 | 0.69 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr10_-_95159933 | 0.68 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr7_-_46569662 | 0.67 |
ENSMUST00000143413.3
ENSMUST00000014546.15 |
Tsg101
|
tumor susceptibility gene 101 |
| chr1_+_138891155 | 0.66 |
ENSMUST00000200533.5
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr6_-_99073156 | 0.66 |
ENSMUST00000175886.8
|
Foxp1
|
forkhead box P1 |
| chr12_+_102249294 | 0.65 |
ENSMUST00000056950.14
|
Rin3
|
Ras and Rab interactor 3 |
| chr9_-_110305705 | 0.65 |
ENSMUST00000198164.5
ENSMUST00000068025.13 |
Klhl18
|
kelch-like 18 |
| chr9_+_102503476 | 0.65 |
ENSMUST00000190279.7
ENSMUST00000188398.7 |
Anapc13
|
anaphase promoting complex subunit 13 |
| chr15_+_35371300 | 0.65 |
ENSMUST00000048646.9
|
Vps13b
|
vacuolar protein sorting 13B |
| chr13_-_74956924 | 0.63 |
ENSMUST00000223206.2
|
Cast
|
calpastatin |
| chr9_+_110306052 | 0.63 |
ENSMUST00000197248.5
ENSMUST00000061155.12 ENSMUST00000198043.5 ENSMUST00000084952.8 |
Kif9
|
kinesin family member 9 |
| chr2_-_168432235 | 0.62 |
ENSMUST00000109184.8
|
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr12_+_76593799 | 0.61 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr3_+_137573436 | 0.59 |
ENSMUST00000090178.10
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr2_+_71884943 | 0.57 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chrX_-_12628309 | 0.56 |
ENSMUST00000096495.11
ENSMUST00000076016.6 |
Med14
|
mediator complex subunit 14 |
| chr7_+_130375799 | 0.56 |
ENSMUST00000048453.7
ENSMUST00000208593.2 |
Btbd16
|
BTB (POZ) domain containing 16 |
| chr5_+_124577952 | 0.56 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr9_+_117869642 | 0.55 |
ENSMUST00000134433.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr19_+_6413703 | 0.55 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr7_+_127475968 | 0.55 |
ENSMUST00000131000.2
|
Zfp646
|
zinc finger protein 646 |
| chr8_-_70448872 | 0.54 |
ENSMUST00000177851.9
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr13_+_41040657 | 0.54 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr8_-_123768759 | 0.53 |
ENSMUST00000098334.13
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr16_-_17540805 | 0.52 |
ENSMUST00000012259.9
ENSMUST00000232236.2 |
Med15
|
mediator complex subunit 15 |
| chr13_-_59970383 | 0.52 |
ENSMUST00000225987.2
|
Tut7
|
terminal uridylyl transferase 7 |
| chr15_-_36609812 | 0.52 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr16_-_17540685 | 0.52 |
ENSMUST00000232163.2
ENSMUST00000232202.2 ENSMUST00000080936.14 ENSMUST00000232645.2 ENSMUST00000232431.2 |
Med15
|
mediator complex subunit 15 |
| chr4_-_156312961 | 0.52 |
ENSMUST00000217885.2
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr15_+_78912642 | 0.51 |
ENSMUST00000180086.3
|
H1f0
|
H1.0 linker histone |
| chr5_-_100563974 | 0.51 |
ENSMUST00000182886.8
ENSMUST00000094578.11 |
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr9_+_57818384 | 0.51 |
ENSMUST00000217129.2
|
Ubl7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chrX_+_109857866 | 0.50 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr14_-_32907446 | 0.50 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr7_+_27879650 | 0.50 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr12_+_102249375 | 0.50 |
ENSMUST00000101114.11
ENSMUST00000150795.8 |
Rin3
|
Ras and Rab interactor 3 |
| chr13_+_41013230 | 0.49 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr2_-_34262012 | 0.49 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr4_-_156312996 | 0.49 |
ENSMUST00000105571.4
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr9_-_57743989 | 0.49 |
ENSMUST00000164010.8
ENSMUST00000171444.8 ENSMUST00000098686.4 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr19_+_6097111 | 0.48 |
ENSMUST00000025723.9
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr4_-_122779837 | 0.48 |
ENSMUST00000106255.8
ENSMUST00000106257.10 |
Cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr19_+_6096606 | 0.48 |
ENSMUST00000138532.8
ENSMUST00000129081.8 ENSMUST00000156550.8 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr4_-_63321591 | 0.48 |
ENSMUST00000035724.5
|
Akna
|
AT-hook transcription factor |
| chr8_-_70449018 | 0.48 |
ENSMUST00000065169.12
ENSMUST00000212478.2 |
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr13_-_59970885 | 0.47 |
ENSMUST00000225179.2
ENSMUST00000225576.2 ENSMUST00000071703.6 |
Tut7
|
terminal uridylyl transferase 7 |
| chr9_+_20556088 | 0.46 |
ENSMUST00000162303.8
ENSMUST00000161486.8 |
Ubl5
|
ubiquitin-like 5 |
| chr3_-_84489923 | 0.46 |
ENSMUST00000143514.3
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr11_-_73742280 | 0.46 |
ENSMUST00000213365.2
|
Olfr393
|
olfactory receptor 393 |
| chr7_+_101619897 | 0.45 |
ENSMUST00000211272.2
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr5_+_117271632 | 0.44 |
ENSMUST00000179276.8
ENSMUST00000092889.12 ENSMUST00000145640.8 |
Taok3
|
TAO kinase 3 |
| chr9_+_20556147 | 0.44 |
ENSMUST00000161882.8
|
Ubl5
|
ubiquitin-like 5 |
| chr17_+_34159633 | 0.44 |
ENSMUST00000025170.11
|
Wdr46
|
WD repeat domain 46 |
| chr4_+_44756608 | 0.43 |
ENSMUST00000143385.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr9_+_117869543 | 0.43 |
ENSMUST00000044454.12
|
Azi2
|
5-azacytidine induced gene 2 |
| chr13_-_104057016 | 0.43 |
ENSMUST00000022222.12
|
Erbin
|
Erbb2 interacting protein |
| chr1_+_39407183 | 0.43 |
ENSMUST00000195123.6
|
Rpl31
|
ribosomal protein L31 |
| chr14_-_98406977 | 0.42 |
ENSMUST00000071533.13
ENSMUST00000069334.8 |
Dach1
|
dachshund family transcription factor 1 |
| chr8_-_64659004 | 0.42 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr17_-_51486196 | 0.42 |
ENSMUST00000024717.10
ENSMUST00000224528.2 |
Tbc1d5
|
TBC1 domain family, member 5 |
| chr1_-_23961379 | 0.41 |
ENSMUST00000027339.14
|
Smap1
|
small ArfGAP 1 |
| chr11_-_98329782 | 0.41 |
ENSMUST00000002655.8
|
Mien1
|
migration and invasion enhancer 1 |
| chr17_+_29487762 | 0.41 |
ENSMUST00000064709.13
ENSMUST00000234711.2 |
BC004004
|
cDNA sequence BC004004 |
| chr11_-_70545450 | 0.40 |
ENSMUST00000018437.3
|
Pfn1
|
profilin 1 |
| chr8_+_72973560 | 0.40 |
ENSMUST00000003123.10
|
Fam32a
|
family with sequence similarity 32, member A |
| chr11_+_69214789 | 0.40 |
ENSMUST00000102602.8
|
Trappc1
|
trafficking protein particle complex 1 |
| chr9_+_117869577 | 0.40 |
ENSMUST00000133580.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr1_-_171910324 | 0.39 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr2_+_145627900 | 0.39 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr11_-_103254257 | 0.39 |
ENSMUST00000092557.6
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr15_-_88703482 | 0.38 |
ENSMUST00000043087.15
ENSMUST00000162183.8 |
Alg12
|
asparagine-linked glycosylation 12 (alpha-1,6-mannosyltransferase) |
| chr2_+_129434738 | 0.38 |
ENSMUST00000153491.8
ENSMUST00000161620.8 ENSMUST00000179001.8 |
Sirpa
|
signal-regulatory protein alpha |
| chr19_+_8897732 | 0.38 |
ENSMUST00000096243.7
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr8_+_72914966 | 0.38 |
ENSMUST00000003121.9
|
Rab8a
|
RAB8A, member RAS oncogene family |
| chr2_-_160714904 | 0.38 |
ENSMUST00000109460.8
ENSMUST00000127201.2 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr9_+_110947167 | 0.38 |
ENSMUST00000035078.13
ENSMUST00000196981.5 ENSMUST00000098340.7 |
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr11_+_117545037 | 0.38 |
ENSMUST00000026658.13
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr2_+_61423469 | 0.38 |
ENSMUST00000112494.2
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr2_+_129434834 | 0.37 |
ENSMUST00000103203.8
|
Sirpa
|
signal-regulatory protein alpha |
| chr8_+_110505494 | 0.37 |
ENSMUST00000034171.9
|
Ap1g1
|
adaptor protein complex AP-1, gamma 1 subunit |
| chr19_-_7183596 | 0.37 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr3_+_96484294 | 0.37 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chr14_+_20724378 | 0.37 |
ENSMUST00000224492.2
ENSMUST00000223751.2 ENSMUST00000225108.2 ENSMUST00000224754.2 |
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr9_+_65816206 | 0.37 |
ENSMUST00000205379.2
ENSMUST00000206048.2 ENSMUST00000034949.10 ENSMUST00000154589.2 |
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr2_+_121287444 | 0.36 |
ENSMUST00000126764.2
|
Hypk
|
huntingtin interacting protein K |
| chr19_+_6097083 | 0.36 |
ENSMUST00000134667.8
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr11_-_115968745 | 0.36 |
ENSMUST00000156545.2
ENSMUST00000075036.9 ENSMUST00000106451.8 |
Unc13d
|
unc-13 homolog D |
| chr7_-_25418401 | 0.36 |
ENSMUST00000125699.3
ENSMUST00000002683.3 |
Ccdc97
|
coiled-coil domain containing 97 |
| chr2_+_61423421 | 0.36 |
ENSMUST00000112495.8
ENSMUST00000112501.9 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr11_-_83189670 | 0.35 |
ENSMUST00000175741.8
ENSMUST00000176518.2 |
Pex12
|
peroxisomal biogenesis factor 12 |
| chr15_-_76079891 | 0.35 |
ENSMUST00000023226.13
|
Plec
|
plectin |
| chr2_+_14078958 | 0.35 |
ENSMUST00000028050.14
|
Stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr7_-_73187369 | 0.35 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr11_-_83189734 | 0.35 |
ENSMUST00000108146.8
ENSMUST00000136369.2 ENSMUST00000018877.9 |
Pex12
|
peroxisomal biogenesis factor 12 |
| chr6_+_29467717 | 0.34 |
ENSMUST00000004396.13
|
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr9_+_27210500 | 0.34 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr11_+_105866030 | 0.33 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr12_+_11316101 | 0.33 |
ENSMUST00000218866.2
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr5_+_122534900 | 0.33 |
ENSMUST00000196969.5
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr11_+_101144533 | 0.33 |
ENSMUST00000007533.15
ENSMUST00000042477.13 ENSMUST00000100414.12 ENSMUST00000107280.11 ENSMUST00000121331.2 |
Vps25
|
vacuolar protein sorting 25 |
| chr11_+_69214971 | 0.33 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chr7_-_28071658 | 0.33 |
ENSMUST00000094644.11
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr13_-_18118736 | 0.33 |
ENSMUST00000009003.9
|
Rala
|
v-ral simian leukemia viral oncogene A (ras related) |
| chr17_-_34159273 | 0.33 |
ENSMUST00000179418.9
ENSMUST00000174048.8 ENSMUST00000174426.2 ENSMUST00000173678.3 ENSMUST00000025163.14 |
Pfdn6
|
prefoldin subunit 6 |
| chr18_+_34994253 | 0.32 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr3_+_27992787 | 0.32 |
ENSMUST00000120834.8
|
Pld1
|
phospholipase D1 |
| chr2_-_92290791 | 0.32 |
ENSMUST00000125276.2
|
Slc35c1
|
solute carrier family 35, member C1 |
| chr11_+_117545618 | 0.32 |
ENSMUST00000106344.8
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr2_-_181333597 | 0.32 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr5_+_143803540 | 0.32 |
ENSMUST00000100487.6
|
Eif2ak1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr5_+_115680964 | 0.32 |
ENSMUST00000137716.8
|
Pxn
|
paxillin |
| chr7_+_126528016 | 0.32 |
ENSMUST00000032924.6
|
Kctd13
|
potassium channel tetramerisation domain containing 13 |
| chr4_-_149569659 | 0.32 |
ENSMUST00000119921.8
|
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr12_-_84408576 | 0.30 |
ENSMUST00000021659.2
ENSMUST00000065536.9 |
Fam161b
|
family with sequence similarity 161, member B |
| chr11_-_4110286 | 0.30 |
ENSMUST00000093381.11
ENSMUST00000101626.9 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr13_-_42000958 | 0.30 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr7_-_30262512 | 0.30 |
ENSMUST00000207747.2
ENSMUST00000207797.2 |
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr17_-_29226886 | 0.29 |
ENSMUST00000232723.2
|
Stk38
|
serine/threonine kinase 38 |
| chr7_-_84339156 | 0.29 |
ENSMUST00000209117.2
ENSMUST00000207975.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr18_-_60981981 | 0.29 |
ENSMUST00000177172.8
ENSMUST00000175934.8 ENSMUST00000176630.8 |
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr5_+_138083345 | 0.29 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr4_+_44756553 | 0.28 |
ENSMUST00000107824.9
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr17_-_57554631 | 0.28 |
ENSMUST00000233568.2
ENSMUST00000005975.8 |
Gpr108
|
G protein-coupled receptor 108 |
| chr2_+_152511381 | 0.28 |
ENSMUST00000125366.8
ENSMUST00000109825.8 ENSMUST00000089059.9 ENSMUST00000079247.4 |
H13
|
histocompatibility 13 |
| chr2_-_73216743 | 0.28 |
ENSMUST00000112044.8
ENSMUST00000112043.8 ENSMUST00000076463.12 |
Gpr155
|
G protein-coupled receptor 155 |
| chr7_-_16020668 | 0.28 |
ENSMUST00000150528.9
ENSMUST00000118976.9 ENSMUST00000146609.3 |
Ccdc9
|
coiled-coil domain containing 9 |
| chr5_-_100126707 | 0.28 |
ENSMUST00000170912.2
|
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr8_-_70962972 | 0.28 |
ENSMUST00000140679.8
ENSMUST00000129909.8 ENSMUST00000081940.11 |
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr14_-_65335479 | 0.27 |
ENSMUST00000225633.2
ENSMUST00000022550.8 |
Extl3
|
exostosin-like glycosyltransferase 3 |
| chr10_-_30531768 | 0.27 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr7_-_6158925 | 0.27 |
ENSMUST00000207957.2
ENSMUST00000094870.3 ENSMUST00000207628.2 |
Zfp787
|
zinc finger protein 787 |
| chr18_+_35860019 | 0.27 |
ENSMUST00000097617.3
|
1700066B19Rik
|
RIKEN cDNA 1700066B19 gene |
| chr11_+_69214883 | 0.27 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr9_+_65816370 | 0.27 |
ENSMUST00000206594.2
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr18_-_46730381 | 0.26 |
ENSMUST00000036030.14
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr18_-_73836810 | 0.26 |
ENSMUST00000025393.14
|
Smad4
|
SMAD family member 4 |
| chrX_-_7956682 | 0.26 |
ENSMUST00000033505.7
|
Was
|
Wiskott-Aldrich syndrome |
| chr2_-_167334746 | 0.25 |
ENSMUST00000109211.9
ENSMUST00000057627.16 |
Spata2
|
spermatogenesis associated 2 |
| chr5_-_69749617 | 0.25 |
ENSMUST00000173927.8
ENSMUST00000120789.8 ENSMUST00000031117.13 |
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr11_+_68393845 | 0.25 |
ENSMUST00000102613.8
ENSMUST00000060441.7 |
Pik3r6
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr1_-_13660027 | 0.25 |
ENSMUST00000027068.11
|
Tram1
|
translocating chain-associating membrane protein 1 |
| chr17_-_36290129 | 0.25 |
ENSMUST00000165613.9
ENSMUST00000173872.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr2_+_25132941 | 0.25 |
ENSMUST00000114355.2
ENSMUST00000060818.2 |
Rnf208
|
ring finger protein 208 |
| chr2_-_26335187 | 0.25 |
ENSMUST00000114082.9
ENSMUST00000091252.5 |
Sec16a
|
SEC16 homolog A, endoplasmic reticulum export factor |
| chr4_-_137084973 | 0.25 |
ENSMUST00000030417.10
ENSMUST00000051477.13 |
Cdc42
|
cell division cycle 42 |
| chr1_+_39406897 | 0.25 |
ENSMUST00000086535.12
|
Rpl31
|
ribosomal protein L31 |
| chr10_+_77458197 | 0.25 |
ENSMUST00000172772.2
|
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chrX_+_41591355 | 0.24 |
ENSMUST00000189753.7
|
Sh2d1a
|
SH2 domain containing 1A |
| chr17_+_32725420 | 0.24 |
ENSMUST00000235238.2
ENSMUST00000165999.2 |
Cyp4f17
|
cytochrome P450, family 4, subfamily f, polypeptide 17 |
| chr17_+_35354430 | 0.24 |
ENSMUST00000173535.8
ENSMUST00000173952.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr7_+_140808680 | 0.24 |
ENSMUST00000106027.9
|
Phrf1
|
PHD and ring finger domains 1 |
| chr4_-_152533265 | 0.24 |
ENSMUST00000159840.8
ENSMUST00000105648.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr18_-_24254865 | 0.24 |
ENSMUST00000055012.12
ENSMUST00000153360.8 ENSMUST00000141489.2 |
Ino80c
|
INO80 complex subunit C |
| chr11_-_58221145 | 0.24 |
ENSMUST00000186859.2
ENSMUST00000065533.3 |
Zfp672
|
zinc finger protein 672 |
| chr11_-_83193412 | 0.24 |
ENSMUST00000176374.2
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr8_-_65582206 | 0.23 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr7_-_141241632 | 0.23 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr18_-_10610332 | 0.23 |
ENSMUST00000025142.13
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr6_+_35229628 | 0.23 |
ENSMUST00000130875.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr8_+_121854566 | 0.23 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr7_-_28071919 | 0.23 |
ENSMUST00000119990.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr19_-_9876815 | 0.23 |
ENSMUST00000237147.2
ENSMUST00000025562.9 |
Incenp
|
inner centromere protein |
| chrX_+_74460234 | 0.23 |
ENSMUST00000033544.14
|
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr15_+_35371644 | 0.23 |
ENSMUST00000227455.2
|
Vps13b
|
vacuolar protein sorting 13B |
| chr2_-_22930149 | 0.22 |
ENSMUST00000091394.13
ENSMUST00000093171.13 |
Abi1
|
abl interactor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spdef
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.3 | 1.4 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.3 | 1.3 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.3 | 0.3 | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) |
| 0.3 | 1.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 0.8 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 1.0 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.2 | 0.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 1.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.5 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.3 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 | 0.3 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 1.0 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 0.5 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.7 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.8 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.3 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) brainstem development(GO:0003360) positive regulation of luteinizing hormone secretion(GO:0033686) nephrogenic mesenchyme development(GO:0072076) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.3 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 2.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.4 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 0.4 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.4 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 | 0.4 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.2 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.3 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.3 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.2 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.1 | 0.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 1.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 1.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.0 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.0 | 0.1 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.9 | GO:0007343 | egg activation(GO:0007343) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 1.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.0 | 0.7 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.3 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.2 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.5 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.5 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 1.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.3 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.3 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.2 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.6 | GO:0033327 | male sex determination(GO:0030238) Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 1.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.7 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.5 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:1903540 | neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.0 | 0.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.5 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.4 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 1.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.0 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 0.9 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.4 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 1.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 2.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.5 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.1 | 0.3 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 1.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 1.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.3 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.2 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 2.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0051425 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.0 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.1 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |