Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
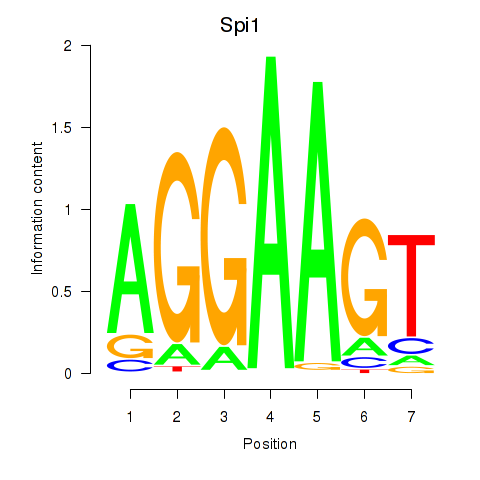
Results for Spi1
Z-value: 1.16
Transcription factors associated with Spi1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spi1
|
ENSMUSG00000002111.9 | spleen focus forming virus (SFFV) proviral integration oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spi1 | mm39_v1_chr2_+_90927053_90927215 | 0.12 | 8.5e-01 | Click! |
Activity profile of Spi1 motif
Sorted Z-values of Spi1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_20296337 | 0.70 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr2_+_84810802 | 0.68 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow |
| chr2_+_84818538 | 0.64 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr3_+_54268523 | 0.52 |
ENSMUST00000117373.8
ENSMUST00000107985.10 ENSMUST00000073012.13 ENSMUST00000081564.13 |
Postn
|
periostin, osteoblast specific factor |
| chr5_-_107873883 | 0.51 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr2_+_129854256 | 0.44 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr4_-_154721288 | 0.36 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr11_-_83540175 | 0.33 |
ENSMUST00000001008.6
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr6_-_52217821 | 0.32 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr7_-_19043955 | 0.30 |
ENSMUST00000207334.2
ENSMUST00000208505.2 ENSMUST00000207716.2 ENSMUST00000208326.2 ENSMUST00000003640.4 |
Fosb
|
FBJ osteosarcoma oncogene B |
| chr9_-_43151179 | 0.30 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr11_-_51748450 | 0.28 |
ENSMUST00000020655.14
ENSMUST00000109090.8 |
Jade2
|
jade family PHD finger 2 |
| chr13_-_23806530 | 0.28 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr11_-_102360664 | 0.28 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr4_+_54947976 | 0.27 |
ENSMUST00000098070.10
|
Zfp462
|
zinc finger protein 462 |
| chr17_+_34809132 | 0.27 |
ENSMUST00000173772.2
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr7_-_99512558 | 0.27 |
ENSMUST00000207137.2
ENSMUST00000207063.2 ENSMUST00000207580.2 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr19_-_33567708 | 0.26 |
ENSMUST00000112508.9
|
Lipo3
|
lipase, member O3 |
| chr18_-_36859732 | 0.26 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr1_-_69723316 | 0.26 |
ENSMUST00000190855.7
ENSMUST00000188110.7 ENSMUST00000191262.7 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr19_+_32597379 | 0.25 |
ENSMUST00000236290.2
ENSMUST00000025833.7 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr2_+_68966125 | 0.25 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chr18_+_61178211 | 0.25 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr14_-_64654397 | 0.24 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr1_+_194621127 | 0.24 |
ENSMUST00000016638.8
ENSMUST00000110815.9 |
Cd34
|
CD34 antigen |
| chr2_+_80145805 | 0.23 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr13_-_110493665 | 0.23 |
ENSMUST00000058806.7
ENSMUST00000224534.2 |
Gapt
|
Grb2-binding adaptor, transmembrane |
| chr19_+_4036562 | 0.23 |
ENSMUST00000236224.2
ENSMUST00000236510.2 ENSMUST00000237910.2 ENSMUST00000235612.2 ENSMUST00000054030.8 |
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr2_+_152468850 | 0.23 |
ENSMUST00000000369.4
ENSMUST00000150913.2 |
Rem1
|
rad and gem related GTP binding protein 1 |
| chr11_+_83594817 | 0.23 |
ENSMUST00000092836.6
|
Wfdc17
|
WAP four-disulfide core domain 17 |
| chr17_+_56312672 | 0.22 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chrM_+_7006 | 0.22 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr5_+_31070739 | 0.22 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr11_-_83469446 | 0.22 |
ENSMUST00000019266.6
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr9_+_66033893 | 0.22 |
ENSMUST00000034945.6
|
Ciao2a
|
cytosolic iron-sulfur assembly component 2A |
| chr18_+_84106188 | 0.21 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr6_+_134012602 | 0.21 |
ENSMUST00000081028.13
ENSMUST00000111963.8 |
Etv6
|
ets variant 6 |
| chr13_-_104057016 | 0.21 |
ENSMUST00000022222.12
|
Erbin
|
Erbb2 interacting protein |
| chr2_+_158148413 | 0.20 |
ENSMUST00000109491.8
ENSMUST00000016168.9 |
Lbp
|
lipopolysaccharide binding protein |
| chr17_+_35200823 | 0.20 |
ENSMUST00000114011.11
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr19_-_33739351 | 0.20 |
ENSMUST00000025694.7
|
Lipo3
|
lipase, member O3 |
| chr7_-_101513300 | 0.20 |
ENSMUST00000106981.8
|
Folr1
|
folate receptor 1 (adult) |
| chr14_-_64654592 | 0.19 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chr4_+_129229805 | 0.19 |
ENSMUST00000119480.2
|
Zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr7_-_101899294 | 0.19 |
ENSMUST00000106923.2
ENSMUST00000098230.11 |
Rhog
|
ras homolog family member G |
| chrX_+_106193167 | 0.19 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr17_+_34808772 | 0.19 |
ENSMUST00000038244.15
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr3_+_95434093 | 0.19 |
ENSMUST00000015667.9
ENSMUST00000116304.3 |
Ctss
|
cathepsin S |
| chr14_-_70396859 | 0.19 |
ENSMUST00000058240.14
ENSMUST00000153871.2 |
9930012K11Rik
|
RIKEN cDNA 9930012K11 gene |
| chr1_+_43484895 | 0.19 |
ENSMUST00000086421.9
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr11_+_68858942 | 0.19 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr6_-_16898440 | 0.18 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr9_+_122752116 | 0.18 |
ENSMUST00000051667.14
|
Zfp105
|
zinc finger protein 105 |
| chr9_+_74945021 | 0.18 |
ENSMUST00000168301.2
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr5_+_52521133 | 0.18 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr2_+_164782642 | 0.18 |
ENSMUST00000137626.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr14_-_79539063 | 0.18 |
ENSMUST00000022595.8
|
Rgcc
|
regulator of cell cycle |
| chr9_-_75448979 | 0.18 |
ENSMUST00000214171.2
|
Tmod3
|
tropomodulin 3 |
| chr5_+_122529941 | 0.18 |
ENSMUST00000102525.11
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr13_+_23922783 | 0.18 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr6_+_4003904 | 0.17 |
ENSMUST00000031670.10
|
Gng11
|
guanine nucleotide binding protein (G protein), gamma 11 |
| chr15_-_66432938 | 0.17 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr6_+_88442391 | 0.17 |
ENSMUST00000032165.16
|
Ruvbl1
|
RuvB-like protein 1 |
| chr17_+_79359617 | 0.17 |
ENSMUST00000233916.2
|
Qpct
|
glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
| chr8_+_95703728 | 0.17 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_+_108811168 | 0.17 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr1_-_131204651 | 0.17 |
ENSMUST00000161764.8
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr1_-_171122509 | 0.17 |
ENSMUST00000111302.4
ENSMUST00000080001.9 |
Ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr17_+_29487881 | 0.17 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr8_+_47986674 | 0.16 |
ENSMUST00000033973.14
ENSMUST00000176379.2 |
Rwdd4a
|
RWD domain containing 4A |
| chr3_-_51184895 | 0.16 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr6_-_136918844 | 0.16 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr9_-_22042930 | 0.16 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr1_+_88022776 | 0.16 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr7_+_105289655 | 0.16 |
ENSMUST00000058333.10
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr19_+_24853039 | 0.16 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr18_+_34994253 | 0.16 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr19_+_4282040 | 0.16 |
ENSMUST00000237518.2
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr6_+_135339543 | 0.16 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr13_-_104056803 | 0.16 |
ENSMUST00000091269.11
ENSMUST00000188997.7 ENSMUST00000169083.8 ENSMUST00000191275.7 |
Erbin
|
Erbb2 interacting protein |
| chr3_-_131138541 | 0.16 |
ENSMUST00000090246.5
ENSMUST00000126569.2 |
Sgms2
|
sphingomyelin synthase 2 |
| chr11_-_98436626 | 0.16 |
ENSMUST00000103141.4
|
Ikzf3
|
IKAROS family zinc finger 3 |
| chr13_-_55676334 | 0.16 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr5_+_102629365 | 0.16 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr5_+_102629240 | 0.16 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr8_-_106022676 | 0.16 |
ENSMUST00000057855.4
|
Exoc3l
|
exocyst complex component 3-like |
| chrX_+_106193060 | 0.16 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr5_-_124003553 | 0.16 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr11_-_78875689 | 0.15 |
ENSMUST00000108269.10
ENSMUST00000108268.10 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr16_-_75706161 | 0.15 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chrX_+_36059274 | 0.15 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr12_+_75355082 | 0.15 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr9_+_58160447 | 0.15 |
ENSMUST00000034883.7
|
Stoml1
|
stomatin-like 1 |
| chr11_+_108316018 | 0.15 |
ENSMUST00000150863.9
ENSMUST00000061287.12 ENSMUST00000149683.9 |
Cep112
|
centrosomal protein 112 |
| chr17_+_47201552 | 0.15 |
ENSMUST00000040434.9
|
Tbcc
|
tubulin-specific chaperone C |
| chr11_-_78875657 | 0.15 |
ENSMUST00000073001.5
|
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr14_-_56339915 | 0.15 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr10_-_111829393 | 0.15 |
ENSMUST00000161870.3
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr11_-_83483807 | 0.15 |
ENSMUST00000019071.4
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr11_-_93859064 | 0.15 |
ENSMUST00000107844.3
ENSMUST00000170303.2 |
Nme1
Gm20390
|
NME/NM23 nucleoside diphosphate kinase 1 predicted gene 20390 |
| chr14_-_52248324 | 0.15 |
ENSMUST00000226964.2
|
Zfp219
|
zinc finger protein 219 |
| chr17_+_43978377 | 0.15 |
ENSMUST00000233627.2
ENSMUST00000233437.2 |
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr3_+_89110223 | 0.15 |
ENSMUST00000077367.11
ENSMUST00000167998.2 |
Gba
|
glucosidase, beta, acid |
| chr9_+_122752164 | 0.15 |
ENSMUST00000148851.2
|
Zfp105
|
zinc finger protein 105 |
| chr15_-_74855264 | 0.14 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr7_+_46445512 | 0.14 |
ENSMUST00000006774.11
ENSMUST00000165031.8 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr11_+_96209093 | 0.14 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chrX_+_133501928 | 0.14 |
ENSMUST00000074950.11
ENSMUST00000113203.2 ENSMUST00000113202.8 ENSMUST00000050331.13 ENSMUST00000059297.6 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chr19_+_53891802 | 0.14 |
ENSMUST00000165617.3
|
Pdcd4
|
programmed cell death 4 |
| chr9_+_107828136 | 0.14 |
ENSMUST00000049348.9
ENSMUST00000194271.2 |
Traip
|
TRAF-interacting protein |
| chr11_-_99276868 | 0.14 |
ENSMUST00000211768.2
|
Krt10
|
keratin 10 |
| chrX_-_69520778 | 0.14 |
ENSMUST00000101506.10
ENSMUST00000114630.3 |
Tmem185a
|
transmembrane protein 185A |
| chr15_-_37007623 | 0.14 |
ENSMUST00000078976.9
|
Zfp706
|
zinc finger protein 706 |
| chr17_+_7437500 | 0.14 |
ENSMUST00000024575.8
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr18_-_35841435 | 0.14 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr3_-_107876477 | 0.14 |
ENSMUST00000004136.10
|
Gstm3
|
glutathione S-transferase, mu 3 |
| chr3_-_106697459 | 0.14 |
ENSMUST00000038845.10
|
Cd53
|
CD53 antigen |
| chr3_+_151143524 | 0.14 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr17_+_27276262 | 0.14 |
ENSMUST00000049308.9
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr8_-_41870077 | 0.14 |
ENSMUST00000033999.8
|
Frg1
|
FSHD region gene 1 |
| chr14_+_56003406 | 0.14 |
ENSMUST00000057569.4
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr17_-_35420972 | 0.14 |
ENSMUST00000167924.2
ENSMUST00000025263.15 |
Tnf
|
tumor necrosis factor |
| chr15_-_63932176 | 0.14 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr6_+_138117519 | 0.14 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr15_+_74828272 | 0.14 |
ENSMUST00000188042.2
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr8_-_72175949 | 0.14 |
ENSMUST00000125092.2
|
Fcho1
|
FCH domain only 1 |
| chr17_+_74796473 | 0.14 |
ENSMUST00000024873.7
|
Yipf4
|
Yip1 domain family, member 4 |
| chr19_+_11747721 | 0.14 |
ENSMUST00000167199.3
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr1_-_71692320 | 0.14 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr17_+_29487762 | 0.13 |
ENSMUST00000064709.13
ENSMUST00000234711.2 |
BC004004
|
cDNA sequence BC004004 |
| chr4_+_133311665 | 0.13 |
ENSMUST00000030661.14
ENSMUST00000105899.2 |
Gpn2
|
GPN-loop GTPase 2 |
| chr19_-_18609118 | 0.13 |
ENSMUST00000025631.7
ENSMUST00000236615.2 |
Ostf1
|
osteoclast stimulating factor 1 |
| chr6_+_124807176 | 0.13 |
ENSMUST00000131847.8
ENSMUST00000151674.3 |
Cdca3
|
cell division cycle associated 3 |
| chr5_+_24598633 | 0.13 |
ENSMUST00000138168.3
ENSMUST00000115077.8 |
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr6_-_136918671 | 0.13 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr6_-_124689001 | 0.13 |
ENSMUST00000203238.2
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr7_+_79910948 | 0.13 |
ENSMUST00000117989.2
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr11_+_98441923 | 0.13 |
ENSMUST00000081033.13
ENSMUST00000107511.8 ENSMUST00000107509.8 ENSMUST00000017339.12 |
Zpbp2
|
zona pellucida binding protein 2 |
| chr6_-_72367642 | 0.13 |
ENSMUST00000059983.10
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr4_-_133599616 | 0.13 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr15_+_78128990 | 0.13 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr9_+_74945101 | 0.13 |
ENSMUST00000167885.8
ENSMUST00000169188.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr5_+_16758538 | 0.13 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor |
| chr11_-_69556904 | 0.13 |
ENSMUST00000018918.12
|
Cd68
|
CD68 antigen |
| chr19_+_32573182 | 0.13 |
ENSMUST00000235594.2
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chrX_+_133486391 | 0.13 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr6_-_67014383 | 0.13 |
ENSMUST00000043098.9
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr7_-_140596811 | 0.13 |
ENSMUST00000081924.5
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr3_-_87676239 | 0.13 |
ENSMUST00000173184.2
ENSMUST00000172621.8 ENSMUST00000174759.8 ENSMUST00000172590.8 ENSMUST00000079083.12 |
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr5_+_16758777 | 0.13 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr2_-_155571279 | 0.13 |
ENSMUST00000040833.5
|
Edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr9_+_72946994 | 0.13 |
ENSMUST00000184126.3
|
Pigbos1
|
Pigb opposite strand 1 |
| chr4_-_133695264 | 0.13 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr18_+_67338437 | 0.13 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr4_+_119052693 | 0.13 |
ENSMUST00000097908.4
|
Svbp
|
small vasohibin binding protein |
| chr8_+_26275560 | 0.13 |
ENSMUST00000211168.2
|
Lsm1
|
LSM1 homolog, mRNA degradation associated |
| chr4_+_45890303 | 0.13 |
ENSMUST00000178561.8
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr6_+_117894242 | 0.13 |
ENSMUST00000180020.8
ENSMUST00000177570.2 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr1_+_152683627 | 0.13 |
ENSMUST00000027754.7
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr9_-_107419309 | 0.13 |
ENSMUST00000195235.6
|
Cyb561d2
|
cytochrome b-561 domain containing 2 |
| chr19_+_4282487 | 0.13 |
ENSMUST00000235306.2
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr8_+_85807566 | 0.13 |
ENSMUST00000140621.2
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr15_-_83054369 | 0.13 |
ENSMUST00000162834.3
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr19_+_6214416 | 0.13 |
ENSMUST00000045042.8
ENSMUST00000237511.2 |
Batf2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr8_-_47986473 | 0.13 |
ENSMUST00000039061.15
|
Trappc11
|
trafficking protein particle complex 11 |
| chr18_-_43610829 | 0.13 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr9_+_74944896 | 0.13 |
ENSMUST00000168166.8
ENSMUST00000169492.8 ENSMUST00000170308.8 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr3_-_58433313 | 0.13 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr8_-_123159663 | 0.12 |
ENSMUST00000017604.10
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr15_-_4008913 | 0.12 |
ENSMUST00000022791.9
|
Fbxo4
|
F-box protein 4 |
| chr1_+_74324089 | 0.12 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr6_+_129022843 | 0.12 |
ENSMUST00000032257.10
ENSMUST00000204320.2 |
Klrb1f
|
killer cell lectin-like receptor subfamily B member 1F |
| chr19_+_53891730 | 0.12 |
ENSMUST00000025931.14
|
Pdcd4
|
programmed cell death 4 |
| chr4_-_43656437 | 0.12 |
ENSMUST00000030192.5
|
Hint2
|
histidine triad nucleotide binding protein 2 |
| chr6_-_40562700 | 0.12 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr1_+_52026296 | 0.12 |
ENSMUST00000168302.8
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr11_-_72441054 | 0.12 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr7_+_44506561 | 0.12 |
ENSMUST00000107876.8
ENSMUST00000154968.2 ENSMUST00000003044.14 |
Pnkp
|
polynucleotide kinase 3'- phosphatase |
| chrX_+_10581248 | 0.12 |
ENSMUST00000144356.8
|
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr9_+_73020708 | 0.12 |
ENSMUST00000169399.8
ENSMUST00000034738.14 |
Rsl24d1
|
ribosomal L24 domain containing 1 |
| chr8_+_95720864 | 0.12 |
ENSMUST00000212141.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr17_+_47816042 | 0.12 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr17_+_43978280 | 0.12 |
ENSMUST00000170988.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr3_-_106126794 | 0.12 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr16_-_56537808 | 0.12 |
ENSMUST00000065515.14
|
Tfg
|
Trk-fused gene |
| chr1_+_178626003 | 0.12 |
ENSMUST00000160789.2
|
Kif26b
|
kinesin family member 26B |
| chr14_+_26722319 | 0.12 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr9_-_108911438 | 0.12 |
ENSMUST00000192801.2
|
Tma7
|
translational machinery associated 7 |
| chr16_+_93404719 | 0.12 |
ENSMUST00000039659.9
ENSMUST00000231762.2 |
Cbr1
|
carbonyl reductase 1 |
| chr9_+_21746785 | 0.12 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr14_-_31503869 | 0.12 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr17_-_48474356 | 0.12 |
ENSMUST00000027764.10
ENSMUST00000053612.14 |
A530064D06Rik
|
RIKEN cDNA A530064D06 gene |
| chr17_-_71297885 | 0.12 |
ENSMUST00000038446.10
|
Myl12b
|
myosin, light chain 12B, regulatory |
| chr5_-_142891565 | 0.12 |
ENSMUST00000171419.8
|
Actb
|
actin, beta |
| chr2_+_90927053 | 0.12 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr3_-_53771185 | 0.12 |
ENSMUST00000122330.2
ENSMUST00000146598.8 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr1_+_152683568 | 0.12 |
ENSMUST00000190323.7
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr5_-_23821523 | 0.12 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr2_-_26823793 | 0.12 |
ENSMUST00000154651.2
ENSMUST00000015011.10 |
Surf4
|
surfeit gene 4 |
| chr4_-_133695204 | 0.12 |
ENSMUST00000100472.10
ENSMUST00000136327.2 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spi1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.2 | 0.7 | GO:0045575 | basophil activation(GO:0045575) |
| 0.1 | 0.6 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.4 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.3 | GO:0060936 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) negative regulation of myofibroblast differentiation(GO:1904761) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.1 | 0.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.4 | GO:0072223 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.1 | 0.2 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.1 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.3 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.2 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.1 | 0.7 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.2 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.3 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.1 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.2 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.2 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.3 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 | 0.3 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.3 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.3 | GO:0038001 | paracrine signaling(GO:0038001) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.2 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:1902022 | L-lysine transport(GO:1902022) |
| 0.0 | 0.2 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.0 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.0 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.0 | GO:0072573 | tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.0 | 0.1 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0071449 | response to lipid hydroperoxide(GO:0006982) cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.1 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.1 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.0 | 0.3 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) zymogen granule exocytosis(GO:0070625) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.0 | 0.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:2000349 | negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.0 | 0.1 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.2 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.3 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.0 | 0.1 | GO:0001805 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0006168 | adenine salvage(GO:0006168) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.2 | GO:0032423 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.0 | 0.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.2 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.0 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.2 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.0 | 0.1 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.0 | 0.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.3 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.0 | GO:0036446 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.2 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0002584 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:2000328 | positive regulation of memory T cell differentiation(GO:0043382) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0046203 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.1 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.1 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.0 | 0.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:1904453 | positive regulation of GTP binding(GO:1904426) regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 | 0.0 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.2 | GO:1902590 | viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.0 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.0 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.1 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.0 | 0.0 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.2 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.0 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 0.0 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.3 | GO:0090662 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.0 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.0 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.1 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0032225 | regulation of synaptic transmission, dopaminergic(GO:0032225) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.0 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.4 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.0 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.0 | GO:0072193 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.0 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.2 | GO:0006544 | glycine metabolic process(GO:0006544) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.2 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0097144 | BAX complex(GO:0097144) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:1990794 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.0 | 0.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.0 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.0 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.7 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.4 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.4 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.3 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.2 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.0 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0003999 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.0 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.0 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 2.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.0 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.0 | GO:0036461 | AP-3 adaptor complex binding(GO:0035651) BLOC-2 complex binding(GO:0036461) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.0 | GO:0051538 | iron-responsive element binding(GO:0030350) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.0 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.0 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 2.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |