Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
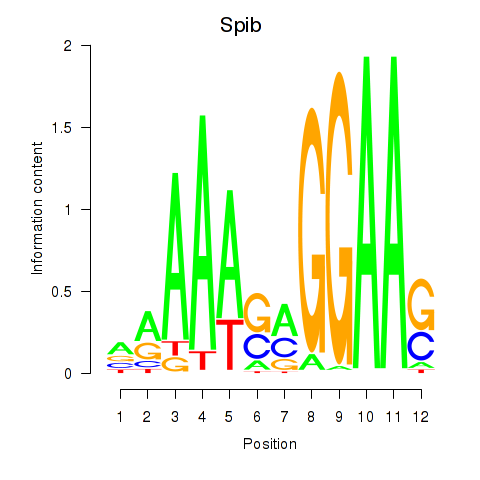
Results for Spib
Z-value: 3.30
Transcription factors associated with Spib
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spib
|
ENSMUSG00000008193.14 | Spi-B transcription factor (Spi-1/PU.1 related) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spib | mm39_v1_chr7_-_44180700_44180805 | -0.52 | 3.7e-01 | Click! |
Activity profile of Spib motif
Sorted Z-values of Spib motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_3918484 | 3.69 |
ENSMUST00000038176.15
ENSMUST00000206077.2 ENSMUST00000090689.5 |
Lilra6
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 |
| chr13_+_23728222 | 3.24 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr3_+_60380243 | 2.72 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr1_+_16758629 | 2.59 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr13_-_23727549 | 2.35 |
ENSMUST00000224359.2
|
H2bc9
|
H2B clustered histone 9 |
| chr7_-_3848050 | 2.27 |
ENSMUST00000108615.10
ENSMUST00000119469.2 |
Pira2
|
paired-Ig-like receptor A2 |
| chr15_+_6552270 | 2.20 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr7_+_3339059 | 2.01 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_-_3723381 | 1.97 |
ENSMUST00000078451.7
|
Pirb
|
paired Ig-like receptor B |
| chr7_+_3339077 | 1.96 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr1_-_171061902 | 1.85 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr7_+_28834276 | 1.85 |
ENSMUST00000161522.8
ENSMUST00000204845.3 ENSMUST00000205027.3 ENSMUST00000204194.3 ENSMUST00000203070.3 ENSMUST00000203380.3 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr6_-_106725929 | 1.74 |
ENSMUST00000204659.3
|
Il5ra
|
interleukin 5 receptor, alpha |
| chr2_-_165996716 | 1.67 |
ENSMUST00000139266.2
|
Sulf2
|
sulfatase 2 |
| chr9_+_106099797 | 1.63 |
ENSMUST00000062241.11
|
Tlr9
|
toll-like receptor 9 |
| chr4_+_132903646 | 1.62 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr7_+_28834350 | 1.62 |
ENSMUST00000159975.8
ENSMUST00000094617.11 ENSMUST00000032811.12 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr5_-_66309244 | 1.55 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr7_+_28834391 | 1.50 |
ENSMUST00000160194.8
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr14_+_54664359 | 1.48 |
ENSMUST00000010550.12
ENSMUST00000199195.3 ENSMUST00000196273.2 |
Mrpl52
|
mitochondrial ribosomal protein L52 |
| chr15_-_78189822 | 1.41 |
ENSMUST00000230115.3
|
Csf2rb2
|
colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) |
| chr10_+_80633865 | 1.40 |
ENSMUST00000148665.8
|
Sf3a2
|
splicing factor 3a, subunit 2 |
| chr3_+_60380463 | 1.37 |
ENSMUST00000195077.6
ENSMUST00000193647.6 ENSMUST00000195001.2 ENSMUST00000192807.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr2_+_145776720 | 1.36 |
ENSMUST00000152515.8
ENSMUST00000138774.8 ENSMUST00000130168.8 ENSMUST00000133433.8 ENSMUST00000118002.2 |
Cfap61
|
cilia and flagella associated protein 61 |
| chr11_+_68322945 | 1.35 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr2_+_22664094 | 1.30 |
ENSMUST00000014290.15
|
Apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr9_-_70411000 | 1.28 |
ENSMUST00000034739.12
|
Rnf111
|
ring finger 111 |
| chr5_+_16758538 | 1.27 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor |
| chr6_+_29529275 | 1.27 |
ENSMUST00000163511.7
|
Irf5
|
interferon regulatory factor 5 |
| chr6_+_122929591 | 1.24 |
ENSMUST00000088468.7
|
Clec4a3
|
C-type lectin domain family 4, member a3 |
| chr18_+_35695736 | 1.23 |
ENSMUST00000235851.2
ENSMUST00000235581.2 |
Matr3
|
matrin 3 |
| chr7_+_75259778 | 1.22 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr11_-_76969230 | 1.21 |
ENSMUST00000102494.8
|
Nsrp1
|
nuclear speckle regulatory protein 1 |
| chr19_-_6134703 | 1.21 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr2_-_181335767 | 1.19 |
ENSMUST00000002532.9
|
Rgs19
|
regulator of G-protein signaling 19 |
| chr11_+_113548201 | 1.17 |
ENSMUST00000148736.8
ENSMUST00000142069.8 ENSMUST00000134418.8 |
Cog1
|
component of oligomeric golgi complex 1 |
| chr2_-_165242307 | 1.17 |
ENSMUST00000029213.5
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr3_-_105839980 | 1.16 |
ENSMUST00000098758.5
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr4_+_62581857 | 1.16 |
ENSMUST00000126338.2
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr15_-_76079891 | 1.14 |
ENSMUST00000023226.13
|
Plec
|
plectin |
| chr2_+_91480513 | 1.10 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr2_-_165997551 | 1.10 |
ENSMUST00000109249.9
ENSMUST00000146497.9 |
Sulf2
|
sulfatase 2 |
| chr5_-_116162415 | 1.08 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr2_-_181335697 | 1.07 |
ENSMUST00000108779.8
ENSMUST00000108769.8 ENSMUST00000108772.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr15_-_78189917 | 1.07 |
ENSMUST00000096356.5
|
Csf2rb2
|
colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) |
| chr9_-_70410611 | 1.06 |
ENSMUST00000215848.2
ENSMUST00000113595.2 ENSMUST00000213647.2 |
Rnf111
|
ring finger 111 |
| chr13_+_20978283 | 1.02 |
ENSMUST00000021757.5
ENSMUST00000221982.2 |
Aoah
|
acyloxyacyl hydrolase |
| chr2_+_113271409 | 1.00 |
ENSMUST00000081349.9
|
Fmn1
|
formin 1 |
| chr7_-_45570828 | 1.00 |
ENSMUST00000038876.13
|
Emp3
|
epithelial membrane protein 3 |
| chr5_+_16758777 | 0.99 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr6_+_137387718 | 0.98 |
ENSMUST00000167002.4
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr6_-_136918671 | 0.97 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr15_+_78210190 | 0.97 |
ENSMUST00000229034.2
ENSMUST00000096355.4 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr5_+_123280250 | 0.97 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr19_+_6413703 | 0.96 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr6_+_137387729 | 0.95 |
ENSMUST00000203914.2
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr8_+_109441276 | 0.95 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr7_-_46569617 | 0.94 |
ENSMUST00000210664.2
ENSMUST00000156335.9 |
Tsg101
|
tumor susceptibility gene 101 |
| chr13_+_30933209 | 0.93 |
ENSMUST00000021784.10
ENSMUST00000110307.3 ENSMUST00000222125.2 |
Irf4
|
interferon regulatory factor 4 |
| chr7_+_127078371 | 0.92 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chr1_+_16758731 | 0.91 |
ENSMUST00000190366.2
|
Ly96
|
lymphocyte antigen 96 |
| chr2_+_91480460 | 0.90 |
ENSMUST00000111331.9
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr16_-_89757693 | 0.90 |
ENSMUST00000002588.11
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr18_+_7869705 | 0.90 |
ENSMUST00000166062.8
ENSMUST00000169010.8 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chr12_+_110413677 | 0.89 |
ENSMUST00000220509.2
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr11_+_69214789 | 0.89 |
ENSMUST00000102602.8
|
Trappc1
|
trafficking protein particle complex 1 |
| chr5_+_16758897 | 0.86 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chr15_+_78210242 | 0.85 |
ENSMUST00000229678.2
ENSMUST00000231888.2 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr7_-_46569662 | 0.84 |
ENSMUST00000143413.3
ENSMUST00000014546.15 |
Tsg101
|
tumor susceptibility gene 101 |
| chr18_+_7869370 | 0.82 |
ENSMUST00000092112.11
ENSMUST00000172018.8 ENSMUST00000168446.8 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chr8_-_32440071 | 0.82 |
ENSMUST00000207678.3
|
Nrg1
|
neuregulin 1 |
| chr1_+_135075377 | 0.81 |
ENSMUST00000125774.2
|
Arl8a
|
ADP-ribosylation factor-like 8A |
| chrX_+_72760183 | 0.80 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr3_+_106628987 | 0.80 |
ENSMUST00000130105.2
|
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr7_-_45546081 | 0.80 |
ENSMUST00000120299.4
ENSMUST00000039049.16 |
Syngr4
|
synaptogyrin 4 |
| chr10_+_51356728 | 0.79 |
ENSMUST00000102894.6
ENSMUST00000219661.2 ENSMUST00000219696.2 ENSMUST00000217706.2 |
Lilr4b
Gm49339
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4B predicted gene, 49339 |
| chr3_+_106629209 | 0.79 |
ENSMUST00000106736.3
ENSMUST00000154973.2 ENSMUST00000098750.5 ENSMUST00000150513.2 |
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr19_+_24976864 | 0.79 |
ENSMUST00000025831.8
|
Dock8
|
dedicator of cytokinesis 8 |
| chr3_-_89959739 | 0.78 |
ENSMUST00000199929.2
ENSMUST00000090908.11 ENSMUST00000198322.5 ENSMUST00000196843.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr2_-_165997179 | 0.77 |
ENSMUST00000088086.4
|
Sulf2
|
sulfatase 2 |
| chr1_-_155912159 | 0.77 |
ENSMUST00000097527.10
|
Tor1aip1
|
torsin A interacting protein 1 |
| chrX_+_99537897 | 0.77 |
ENSMUST00000033570.6
|
Igbp1
|
immunoglobulin (CD79A) binding protein 1 |
| chr2_+_145776697 | 0.77 |
ENSMUST00000116398.8
ENSMUST00000126415.8 |
Cfap61
|
cilia and flagella associated protein 61 |
| chr2_-_181333597 | 0.77 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr3_-_89959770 | 0.77 |
ENSMUST00000029553.16
ENSMUST00000195995.5 ENSMUST00000064639.15 ENSMUST00000199834.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr2_+_24235300 | 0.76 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr12_+_33365371 | 0.75 |
ENSMUST00000154742.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr11_+_69214971 | 0.75 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chr5_-_102217770 | 0.75 |
ENSMUST00000053177.14
ENSMUST00000174698.2 |
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr12_-_65012270 | 0.74 |
ENSMUST00000222508.2
|
Klhl28
|
kelch-like 28 |
| chr3_-_129854477 | 0.73 |
ENSMUST00000001079.15
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr13_+_9143916 | 0.72 |
ENSMUST00000188211.8
ENSMUST00000188939.7 ENSMUST00000190041.7 |
Larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr13_+_110039620 | 0.71 |
ENSMUST00000120664.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr6_-_124710030 | 0.70 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr15_-_78456898 | 0.69 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr2_-_181335518 | 0.68 |
ENSMUST00000108776.8
ENSMUST00000108771.2 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr11_+_11635908 | 0.67 |
ENSMUST00000065433.12
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr9_-_114811807 | 0.67 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr2_+_32611067 | 0.67 |
ENSMUST00000074248.11
|
Sh2d3c
|
SH2 domain containing 3C |
| chr7_-_44888532 | 0.66 |
ENSMUST00000033063.15
|
Cd37
|
CD37 antigen |
| chr19_+_26600820 | 0.66 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_10085299 | 0.66 |
ENSMUST00000114897.9
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr9_+_117869642 | 0.66 |
ENSMUST00000134433.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr8_+_3403415 | 0.65 |
ENSMUST00000098966.5
ENSMUST00000239102.2 |
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18 |
| chr2_+_90927053 | 0.64 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr11_-_34674677 | 0.63 |
ENSMUST00000093193.12
ENSMUST00000101365.9 |
Dock2
|
dedicator of cyto-kinesis 2 |
| chr14_+_80237691 | 0.62 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr2_-_10085133 | 0.62 |
ENSMUST00000145530.8
ENSMUST00000026887.14 ENSMUST00000114896.8 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr11_+_69214883 | 0.62 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr16_-_92622972 | 0.61 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr2_+_126549239 | 0.61 |
ENSMUST00000028841.14
ENSMUST00000110416.3 |
Usp8
|
ubiquitin specific peptidase 8 |
| chr15_-_63932176 | 0.60 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr1_+_155911518 | 0.59 |
ENSMUST00000133152.2
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr1_-_170886924 | 0.58 |
ENSMUST00000164044.8
ENSMUST00000169017.8 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr7_-_3298243 | 0.58 |
ENSMUST00000108653.4
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr19_+_6107874 | 0.57 |
ENSMUST00000178310.9
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr15_-_82917495 | 0.56 |
ENSMUST00000231165.2
|
Nfam1
|
Nfat activating molecule with ITAM motif 1 |
| chr18_+_75133519 | 0.56 |
ENSMUST00000079716.6
|
Rpl17
|
ribosomal protein L17 |
| chr6_-_113354826 | 0.56 |
ENSMUST00000032410.14
|
Tada3
|
transcriptional adaptor 3 |
| chr2_-_45001141 | 0.56 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr13_-_13568106 | 0.55 |
ENSMUST00000021738.10
ENSMUST00000220628.2 |
Gpr137b
|
G protein-coupled receptor 137B |
| chr11_-_58221145 | 0.55 |
ENSMUST00000186859.2
ENSMUST00000065533.3 |
Zfp672
|
zinc finger protein 672 |
| chr8_+_111345209 | 0.55 |
ENSMUST00000034190.11
|
Vac14
|
Vac14 homolog (S. cerevisiae) |
| chr5_-_138262178 | 0.55 |
ENSMUST00000048421.14
|
Map11
|
microtubule associated protein 11 |
| chr13_-_103470937 | 0.54 |
ENSMUST00000167058.8
ENSMUST00000164111.2 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr9_+_64718708 | 0.54 |
ENSMUST00000213926.2
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr16_-_22475915 | 0.53 |
ENSMUST00000089925.10
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr12_-_28685913 | 0.52 |
ENSMUST00000074267.5
|
Rps7
|
ribosomal protein S7 |
| chr2_+_112096154 | 0.52 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr18_-_39622932 | 0.51 |
ENSMUST00000152853.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr16_-_56537808 | 0.49 |
ENSMUST00000065515.14
|
Tfg
|
Trk-fused gene |
| chr10_+_81229443 | 0.49 |
ENSMUST00000118206.2
|
Smim24
|
small integral membrane protein 24 |
| chr13_+_102830029 | 0.49 |
ENSMUST00000022124.10
ENSMUST00000171267.2 ENSMUST00000167144.2 ENSMUST00000170878.2 |
Cd180
|
CD180 antigen |
| chr19_+_4281953 | 0.49 |
ENSMUST00000025773.5
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr4_-_154245073 | 0.49 |
ENSMUST00000105639.4
ENSMUST00000030896.15 |
Tprgl
|
transformation related protein 63 regulated like |
| chr9_+_100956145 | 0.49 |
ENSMUST00000189616.2
|
Msl2
|
MSL complex subunit 2 |
| chrX_-_37545311 | 0.48 |
ENSMUST00000074913.12
ENSMUST00000016678.14 ENSMUST00000061755.9 |
Lamp2
|
lysosomal-associated membrane protein 2 |
| chr11_+_69214895 | 0.47 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chr12_-_84408576 | 0.47 |
ENSMUST00000021659.2
ENSMUST00000065536.9 |
Fam161b
|
family with sequence similarity 161, member B |
| chr4_+_86666764 | 0.47 |
ENSMUST00000045512.15
ENSMUST00000082026.14 |
Dennd4c
|
DENN/MADD domain containing 4C |
| chr16_-_22475960 | 0.47 |
ENSMUST00000023578.14
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr19_-_6134903 | 0.47 |
ENSMUST00000160977.8
ENSMUST00000159859.2 ENSMUST00000025707.9 ENSMUST00000160712.8 ENSMUST00000237738.2 |
Zfpl1
|
zinc finger like protein 1 |
| chr5_-_25703700 | 0.47 |
ENSMUST00000173073.8
ENSMUST00000045291.14 ENSMUST00000173174.2 |
Kmt2c
|
lysine (K)-specific methyltransferase 2C |
| chr6_-_106725895 | 0.46 |
ENSMUST00000205004.2
|
Il5ra
|
interleukin 5 receptor, alpha |
| chr9_+_117869543 | 0.46 |
ENSMUST00000044454.12
|
Azi2
|
5-azacytidine induced gene 2 |
| chr18_-_35348049 | 0.46 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr7_+_43057611 | 0.46 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr4_-_95965767 | 0.46 |
ENSMUST00000030305.13
ENSMUST00000107078.9 |
Cyp2j13
|
cytochrome P450, family 2, subfamily j, polypeptide 13 |
| chr16_-_36486429 | 0.45 |
ENSMUST00000089620.11
|
Cd86
|
CD86 antigen |
| chr11_-_103235475 | 0.45 |
ENSMUST00000041385.14
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr9_+_123901979 | 0.44 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr2_+_43445333 | 0.43 |
ENSMUST00000028223.9
ENSMUST00000112826.8 |
Kynu
|
kynureninase |
| chr9_+_117869577 | 0.43 |
ENSMUST00000133580.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr13_+_81034214 | 0.42 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr7_-_28135616 | 0.42 |
ENSMUST00000208199.2
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr8_-_123302187 | 0.42 |
ENSMUST00000213062.2
|
Aprt
|
adenine phosphoribosyl transferase |
| chr5_+_115680964 | 0.42 |
ENSMUST00000137716.8
|
Pxn
|
paxillin |
| chr9_-_37259629 | 0.41 |
ENSMUST00000217238.3
|
Ccdc15
|
coiled-coil domain containing 15 |
| chr4_-_43454600 | 0.40 |
ENSMUST00000098105.4
ENSMUST00000098104.10 ENSMUST00000030179.11 |
Cd72
|
CD72 antigen |
| chr4_-_118266416 | 0.40 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr15_-_36609208 | 0.40 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_-_101831729 | 0.39 |
ENSMUST00000190824.7
|
Slc22a15
|
solute carrier family 22 (organic anion/cation transporter), member 15 |
| chr6_+_129326927 | 0.39 |
ENSMUST00000065289.6
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr13_+_20369377 | 0.39 |
ENSMUST00000221067.2
|
Elmo1
|
engulfment and cell motility 1 |
| chr19_-_8751795 | 0.39 |
ENSMUST00000010249.7
|
Tmem179b
|
transmembrane protein 179B |
| chr2_-_77776524 | 0.38 |
ENSMUST00000111824.8
ENSMUST00000111819.8 ENSMUST00000128963.2 |
Cwc22
|
CWC22 spliceosome-associated protein |
| chr2_+_72115981 | 0.38 |
ENSMUST00000090824.12
ENSMUST00000135469.8 |
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr8_+_84262409 | 0.38 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr4_-_122779837 | 0.38 |
ENSMUST00000106255.8
ENSMUST00000106257.10 |
Cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr13_-_53627110 | 0.37 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr14_+_66872699 | 0.37 |
ENSMUST00000159365.8
ENSMUST00000054661.8 ENSMUST00000225182.2 ENSMUST00000159068.2 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr3_-_79439181 | 0.36 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2 |
| chr15_+_97259060 | 0.35 |
ENSMUST00000228521.2
ENSMUST00000226495.2 |
Pced1b
|
PC-esterase domain containing 1B |
| chr5_-_32903687 | 0.35 |
ENSMUST00000135248.2
|
Pisd
|
phosphatidylserine decarboxylase |
| chr7_+_135207505 | 0.35 |
ENSMUST00000210833.2
|
Ptpre
|
protein tyrosine phosphatase, receptor type, E |
| chr13_+_102830104 | 0.34 |
ENSMUST00000172138.2
|
Cd180
|
CD180 antigen |
| chr1_-_80191649 | 0.34 |
ENSMUST00000058748.2
|
Fam124b
|
family with sequence similarity 124, member B |
| chr13_-_23650045 | 0.34 |
ENSMUST00000041674.14
ENSMUST00000110434.2 |
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr13_+_44884740 | 0.34 |
ENSMUST00000173246.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr15_-_9529898 | 0.33 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr17_-_47015928 | 0.33 |
ENSMUST00000002839.9
ENSMUST00000233988.2 |
Ppp2r5d
|
protein phosphatase 2, regulatory subunit B', delta |
| chr12_+_98234884 | 0.33 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr10_+_128584324 | 0.32 |
ENSMUST00000065210.10
ENSMUST00000218218.2 |
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr12_-_32258604 | 0.31 |
ENSMUST00000053215.14
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr3_-_89959917 | 0.31 |
ENSMUST00000197903.5
|
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr4_+_44012638 | 0.30 |
ENSMUST00000107847.10
ENSMUST00000170241.8 ENSMUST00000107849.10 ENSMUST00000107851.10 ENSMUST00000107845.4 |
Clta
|
clathrin, light polypeptide (Lca) |
| chr17_+_21165573 | 0.30 |
ENSMUST00000007708.14
|
Ppp2r1a
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr12_+_77285770 | 0.30 |
ENSMUST00000062804.8
|
Fut8
|
fucosyltransferase 8 |
| chr1_-_38704028 | 0.30 |
ENSMUST00000039827.14
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr6_-_30304512 | 0.30 |
ENSMUST00000094543.3
ENSMUST00000102993.10 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr3_-_89294430 | 0.30 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr19_+_18609291 | 0.29 |
ENSMUST00000042392.14
ENSMUST00000237347.2 |
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr2_-_91480096 | 0.28 |
ENSMUST00000099714.10
ENSMUST00000111333.2 |
Zfp408
|
zinc finger protein 408 |
| chr17_-_56891576 | 0.27 |
ENSMUST00000075510.12
|
Safb2
|
scaffold attachment factor B2 |
| chr2_-_91014163 | 0.27 |
ENSMUST00000077941.13
ENSMUST00000111381.9 ENSMUST00000111372.8 ENSMUST00000111371.8 ENSMUST00000075269.10 ENSMUST00000066473.12 |
Madd
|
MAP-kinase activating death domain |
| chr1_+_87548026 | 0.27 |
ENSMUST00000169754.8
ENSMUST00000042275.15 ENSMUST00000168783.8 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr10_+_78848306 | 0.27 |
ENSMUST00000216030.2
|
Olfr1351
|
olfactory receptor 1351 |
| chr2_+_43445359 | 0.27 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr13_+_9143995 | 0.26 |
ENSMUST00000091829.4
|
Larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr6_-_136918495 | 0.26 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spib
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.5 | 1.6 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.5 | 3.5 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.5 | 4.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 1.9 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.5 | 1.9 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.4 | 1.8 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.3 | 1.7 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.3 | 2.0 | GO:1903336 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 0.3 | 0.9 | GO:0045404 | interleukin-10 biosynthetic process(GO:0042091) interleukin-13 biosynthetic process(GO:0042231) regulation of interleukin-10 biosynthetic process(GO:0045074) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.3 | 1.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 0.9 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.3 | 1.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 3.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 0.5 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.2 | 0.7 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.2 | 2.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 0.9 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 0.7 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.2 | 0.7 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.2 | 0.6 | GO:0090264 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.2 | 0.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 0.9 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.7 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 1.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 0.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 1.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.2 | 0.7 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) positive regulation of progesterone secretion(GO:2000872) |
| 0.2 | 0.8 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 1.2 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 1.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.4 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 1.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.4 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 1.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.4 | GO:0006168 | adenine salvage(GO:0006168) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.9 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.3 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.5 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 0.7 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.4 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.3 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 1.3 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 4.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.7 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.8 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 2.2 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.1 | 0.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 | 0.2 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.1 | 0.3 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.4 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 0.2 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 4.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 1.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.8 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 1.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.3 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 1.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 1.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 1.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.7 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 2.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 1.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.4 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 1.0 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.3 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.9 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.4 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.9 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.2 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.3 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 1.3 | GO:0045727 | positive regulation of translation(GO:0045727) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.4 | 3.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 2.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 1.6 | GO:0036019 | endolysosome(GO:0036019) |
| 0.2 | 0.7 | GO:0097635 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.2 | 1.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0097637 | platelet dense granule membrane(GO:0031088) intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 2.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.7 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 2.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 4.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 3.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.5 | 3.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 1.8 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.3 | 4.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 2.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 1.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 1.0 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 0.9 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 0.6 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.2 | 0.8 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.2 | 0.7 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.7 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.1 | 0.4 | GO:0003999 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.1 | 1.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.7 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.1 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 0.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 3.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 4.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 3.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 7.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 1.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 1.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.4 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 4.3 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 3.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 3.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 2.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 3.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 3.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 3.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 3.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.5 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 1.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 3.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 2.5 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |