Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
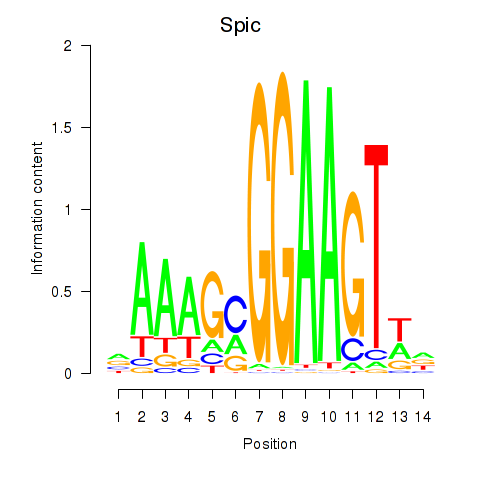
Results for Spic
Z-value: 0.46
Transcription factors associated with Spic
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spic
|
ENSMUSG00000004359.17 | Spi-C transcription factor (Spi-1/PU.1 related) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spic | mm39_v1_chr10_-_88518878_88518892 | -0.51 | 3.8e-01 | Click! |
Activity profile of Spic motif
Sorted Z-values of Spic motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_80145805 | 0.54 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr9_+_20556147 | 0.29 |
ENSMUST00000161882.8
|
Ubl5
|
ubiquitin-like 5 |
| chr4_+_129229805 | 0.28 |
ENSMUST00000119480.2
|
Zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr11_+_87684548 | 0.28 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr8_+_95650315 | 0.27 |
ENSMUST00000153448.9
ENSMUST00000166802.9 ENSMUST00000074570.10 |
Adgrg5
|
adhesion G protein-coupled receptor G5 |
| chr19_-_41884599 | 0.26 |
ENSMUST00000038677.5
|
Rrp12
|
ribosomal RNA processing 12 homolog |
| chr11_+_87684299 | 0.26 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr7_-_126303887 | 0.25 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr19_+_4282040 | 0.23 |
ENSMUST00000237518.2
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr6_-_56681657 | 0.22 |
ENSMUST00000176595.3
ENSMUST00000170382.5 ENSMUST00000203958.2 |
Lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_-_94794256 | 0.22 |
ENSMUST00000005923.7
|
Psmb4
|
proteasome (prosome, macropain) subunit, beta type 4 |
| chr7_-_126303689 | 0.20 |
ENSMUST00000135087.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr5_-_138169253 | 0.19 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_138169509 | 0.19 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr4_-_136620376 | 0.19 |
ENSMUST00000046332.6
|
C1qc
|
complement component 1, q subcomponent, C chain |
| chr17_+_75024727 | 0.18 |
ENSMUST00000024882.8
ENSMUST00000234751.2 ENSMUST00000234568.2 |
Ttc27
|
tetratricopeptide repeat domain 27 |
| chr17_+_13135313 | 0.18 |
ENSMUST00000143961.2
ENSMUST00000233286.2 |
Tcp1
|
t-complex protein 1 |
| chr3_-_65299967 | 0.18 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr10_-_17898838 | 0.17 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr4_-_117749191 | 0.17 |
ENSMUST00000030265.4
|
Dph2
|
DPH2 homolog |
| chr12_+_70499869 | 0.16 |
ENSMUST00000021471.13
|
Tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr8_+_107783502 | 0.16 |
ENSMUST00000034392.13
ENSMUST00000170962.3 |
Nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr19_+_11747721 | 0.16 |
ENSMUST00000167199.3
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr10_-_116899239 | 0.16 |
ENSMUST00000219036.2
ENSMUST00000218059.2 |
Cct2
|
chaperonin containing Tcp1, subunit 2 (beta) |
| chr13_-_58276353 | 0.16 |
ENSMUST00000007980.7
|
Hnrnpa0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr4_-_136626073 | 0.16 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr2_+_43638814 | 0.15 |
ENSMUST00000112824.8
ENSMUST00000055776.8 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr6_-_136918844 | 0.15 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chrX_-_168103266 | 0.15 |
ENSMUST00000033717.9
ENSMUST00000112115.2 |
Hccs
|
holocytochrome c synthetase |
| chr5_+_149202157 | 0.15 |
ENSMUST00000200806.4
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr7_-_44888465 | 0.15 |
ENSMUST00000210078.2
|
Cd37
|
CD37 antigen |
| chr10_+_79715448 | 0.15 |
ENSMUST00000006679.15
|
Prtn3
|
proteinase 3 |
| chr10_-_17898977 | 0.15 |
ENSMUST00000020002.9
|
Abracl
|
ABRA C-terminal like |
| chr1_+_152683568 | 0.15 |
ENSMUST00000190323.7
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr14_+_8507874 | 0.15 |
ENSMUST00000225325.2
|
Thoc7
|
THO complex 7 |
| chrX_+_7750558 | 0.15 |
ENSMUST00000208640.2
ENSMUST00000207114.2 ENSMUST00000208633.2 ENSMUST00000208397.2 ENSMUST00000153620.3 ENSMUST00000123277.8 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr10_-_116899664 | 0.15 |
ENSMUST00000218719.2
ENSMUST00000219573.2 ENSMUST00000047672.9 |
Cct2
|
chaperonin containing Tcp1, subunit 2 (beta) |
| chr12_+_100076407 | 0.15 |
ENSMUST00000021595.10
|
Psmc1
|
protease (prosome, macropain) 26S subunit, ATPase 1 |
| chr10_-_17898938 | 0.14 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chrX_-_50106844 | 0.14 |
ENSMUST00000053593.8
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr17_-_35391583 | 0.14 |
ENSMUST00000173106.2
|
Aif1
|
allograft inflammatory factor 1 |
| chr2_+_163836880 | 0.14 |
ENSMUST00000018470.10
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
| chr12_+_75355082 | 0.14 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr7_-_19005721 | 0.13 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr4_-_136613498 | 0.13 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr11_-_69553390 | 0.13 |
ENSMUST00000129224.8
ENSMUST00000155200.8 |
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr1_-_149836974 | 0.13 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chrX_-_133442596 | 0.13 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr11_+_78215026 | 0.13 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr11_-_119190830 | 0.13 |
ENSMUST00000106253.2
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr3_-_108053396 | 0.13 |
ENSMUST00000000001.5
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr11_+_58221538 | 0.13 |
ENSMUST00000116376.9
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr5_-_140368482 | 0.13 |
ENSMUST00000196566.5
|
Snx8
|
sorting nexin 8 |
| chr9_+_107440484 | 0.13 |
ENSMUST00000193418.2
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr11_-_113456568 | 0.13 |
ENSMUST00000071539.10
ENSMUST00000106633.10 ENSMUST00000042657.16 ENSMUST00000149034.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr13_-_96807346 | 0.12 |
ENSMUST00000022176.15
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr9_-_114610879 | 0.12 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr9_-_105398346 | 0.12 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr12_+_84408742 | 0.12 |
ENSMUST00000021661.13
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr8_-_85807281 | 0.11 |
ENSMUST00000152785.8
|
Wdr83
|
WD repeat domain containing 83 |
| chr6_-_124710030 | 0.11 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_-_65300000 | 0.11 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr19_+_6135013 | 0.11 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr4_+_140428777 | 0.11 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr11_+_70021155 | 0.11 |
ENSMUST00000041550.12
ENSMUST00000165951.8 |
Mgl2
|
macrophage galactose N-acetyl-galactosamine specific lectin 2 |
| chrX_-_135642025 | 0.11 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr9_+_7184514 | 0.11 |
ENSMUST00000215683.2
ENSMUST00000034499.10 |
Dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr9_+_46184362 | 0.11 |
ENSMUST00000156440.8
ENSMUST00000114552.4 |
Zpr1
|
ZPR1 zinc finger |
| chr12_+_55445560 | 0.11 |
ENSMUST00000021412.9
|
Psma6
|
proteasome subunit alpha 6 |
| chr1_-_170804116 | 0.10 |
ENSMUST00000159969.8
|
Fcgr2b
|
Fc receptor, IgG, low affinity IIb |
| chr19_-_6886965 | 0.10 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr8_+_93581946 | 0.10 |
ENSMUST00000046290.3
ENSMUST00000210099.2 |
Lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr15_+_76253084 | 0.10 |
ENSMUST00000023213.8
|
Hgh1
|
HGH1 homolog |
| chr11_+_58221569 | 0.10 |
ENSMUST00000073128.7
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chrX_+_7750483 | 0.10 |
ENSMUST00000115663.10
ENSMUST00000096514.11 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr14_-_56026266 | 0.10 |
ENSMUST00000168716.8
ENSMUST00000178399.3 ENSMUST00000022830.14 |
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr9_+_56858162 | 0.10 |
ENSMUST00000068856.5
|
Snupn
|
snurportin 1 |
| chr17_-_6923299 | 0.10 |
ENSMUST00000179554.3
|
Dynlt1f
|
dynein light chain Tctex-type 1F |
| chr17_+_13135216 | 0.10 |
ENSMUST00000089024.13
ENSMUST00000151287.8 |
Tcp1
|
t-complex protein 1 |
| chr11_-_69873343 | 0.10 |
ENSMUST00000147437.3
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr8_-_70169500 | 0.10 |
ENSMUST00000238408.2
ENSMUST00000238418.2 ENSMUST00000238753.2 |
Zfp869
|
zinc finger protein 869 |
| chr16_-_75563645 | 0.10 |
ENSMUST00000114244.2
ENSMUST00000046283.16 |
Hspa13
|
heat shock protein 70 family, member 13 |
| chr8_+_22682816 | 0.10 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr8_-_85807308 | 0.10 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr3_-_106697459 | 0.10 |
ENSMUST00000038845.10
|
Cd53
|
CD53 antigen |
| chr8_+_85798348 | 0.10 |
ENSMUST00000078665.13
|
Dhps
|
deoxyhypusine synthase |
| chr6_+_129326927 | 0.10 |
ENSMUST00000065289.6
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr9_+_109704609 | 0.09 |
ENSMUST00000094324.8
|
Cdc25a
|
cell division cycle 25A |
| chr5_-_110928436 | 0.09 |
ENSMUST00000149208.2
ENSMUST00000031483.15 ENSMUST00000086643.12 ENSMUST00000170468.8 ENSMUST00000031481.13 |
Pus1
|
pseudouridine synthase 1 |
| chr16_+_32238520 | 0.09 |
ENSMUST00000014220.15
ENSMUST00000080316.8 |
Dynlt2b
|
dynein light chain Tctex-type 2B |
| chr5_-_110987441 | 0.09 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chrX_+_7750261 | 0.09 |
ENSMUST00000115660.12
|
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr11_-_119190896 | 0.09 |
ENSMUST00000026667.15
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr19_+_6111204 | 0.09 |
ENSMUST00000162726.5
|
Znhit2
|
zinc finger, HIT domain containing 2 |
| chr6_+_113581708 | 0.09 |
ENSMUST00000035725.7
|
Brk1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr6_+_134391385 | 0.09 |
ENSMUST00000111960.2
|
Bcl2l14
|
BCL2-like 14 (apoptosis facilitator) |
| chr17_-_24292453 | 0.09 |
ENSMUST00000017090.6
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr13_+_90237824 | 0.09 |
ENSMUST00000012566.9
|
Tmem167
|
transmembrane protein 167 |
| chr17_+_6869070 | 0.09 |
ENSMUST00000092966.5
|
Dynlt1c
|
dynein light chain Tctex-type 1C |
| chr6_-_48685108 | 0.09 |
ENSMUST00000126422.3
ENSMUST00000119315.2 ENSMUST00000053661.7 |
Gimap6
|
GTPase, IMAP family member 6 |
| chr15_+_78314251 | 0.09 |
ENSMUST00000229622.2
ENSMUST00000162808.2 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr3_+_135191366 | 0.09 |
ENSMUST00000029814.10
|
Manba
|
mannosidase, beta A, lysosomal |
| chr4_-_41314877 | 0.09 |
ENSMUST00000030145.9
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr7_+_127368981 | 0.09 |
ENSMUST00000118865.8
ENSMUST00000061587.13 ENSMUST00000121504.2 |
Orai3
|
ORAI calcium release-activated calcium modulator 3 |
| chr4_-_117744476 | 0.08 |
ENSMUST00000132073.2
ENSMUST00000150204.8 ENSMUST00000147845.2 ENSMUST00000036380.14 ENSMUST00000136596.2 |
Atp6v0b
|
ATPase, H+ transporting, lysosomal V0 subunit B |
| chr11_-_102109748 | 0.08 |
ENSMUST00000131254.2
|
Hdac5
|
histone deacetylase 5 |
| chr9_+_99457829 | 0.08 |
ENSMUST00000066650.12
ENSMUST00000148987.8 |
Dbr1
|
debranching RNA lariats 1 |
| chr12_-_80690573 | 0.08 |
ENSMUST00000166931.2
ENSMUST00000218364.2 |
Erh
|
ERH mRNA splicing and mitosis factor |
| chr5_+_115465205 | 0.08 |
ENSMUST00000031513.14
|
Srsf9
|
serine and arginine-rich splicing factor 9 |
| chr1_+_52158721 | 0.08 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr2_+_130266253 | 0.08 |
ENSMUST00000128994.8
ENSMUST00000028900.11 |
Vps16
|
VSP16 CORVET/HOPS core subunit |
| chr15_-_63932176 | 0.08 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr15_+_103362195 | 0.08 |
ENSMUST00000047405.9
|
Nckap1l
|
NCK associated protein 1 like |
| chr13_-_39144475 | 0.08 |
ENSMUST00000225331.2
ENSMUST00000224645.2 ENSMUST00000167513.3 ENSMUST00000225568.2 |
Slc35b3
|
solute carrier family 35, member B3 |
| chr1_-_171122509 | 0.08 |
ENSMUST00000111302.4
ENSMUST00000080001.9 |
Ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr5_+_149201577 | 0.08 |
ENSMUST00000071130.5
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr9_-_37568570 | 0.08 |
ENSMUST00000117654.3
|
Tbrg1
|
transforming growth factor beta regulated gene 1 |
| chr9_+_108911547 | 0.08 |
ENSMUST00000026735.9
|
Ccdc51
|
coiled-coil domain containing 51 |
| chr8_+_126456710 | 0.08 |
ENSMUST00000143504.8
|
Ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr7_+_28533279 | 0.08 |
ENSMUST00000208971.2
ENSMUST00000066723.15 |
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr11_-_102771751 | 0.08 |
ENSMUST00000021306.14
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr5_+_86219593 | 0.08 |
ENSMUST00000198435.5
ENSMUST00000031171.9 |
Stap1
|
signal transducing adaptor family member 1 |
| chr7_+_28140352 | 0.08 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr6_+_123206802 | 0.08 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr11_+_106146966 | 0.08 |
ENSMUST00000021049.9
|
Psmc5
|
protease (prosome, macropain) 26S subunit, ATPase 5 |
| chr7_+_44506731 | 0.08 |
ENSMUST00000098478.7
|
Pnkp
|
polynucleotide kinase 3'- phosphatase |
| chr13_-_98399533 | 0.08 |
ENSMUST00000040972.4
|
Utp15
|
UTP15 small subunit processome component |
| chr15_-_57755753 | 0.08 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr19_+_12438125 | 0.07 |
ENSMUST00000081035.9
|
Mpeg1
|
macrophage expressed gene 1 |
| chr11_+_88095222 | 0.07 |
ENSMUST00000118784.8
ENSMUST00000139170.8 ENSMUST00000107915.10 ENSMUST00000144070.3 |
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr7_+_105289655 | 0.07 |
ENSMUST00000058333.10
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr7_+_44506561 | 0.07 |
ENSMUST00000107876.8
ENSMUST00000154968.2 ENSMUST00000003044.14 |
Pnkp
|
polynucleotide kinase 3'- phosphatase |
| chr18_+_67338437 | 0.07 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr4_-_133601990 | 0.07 |
ENSMUST00000168974.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr7_-_126074222 | 0.07 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr2_-_25101354 | 0.07 |
ENSMUST00000059849.15
|
Nelfb
|
negative elongation factor complex member B |
| chr11_-_106605772 | 0.07 |
ENSMUST00000124958.3
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr19_-_18609118 | 0.07 |
ENSMUST00000025631.7
ENSMUST00000236615.2 |
Ostf1
|
osteoclast stimulating factor 1 |
| chr12_+_84408803 | 0.07 |
ENSMUST00000110278.8
ENSMUST00000145522.2 |
Coq6
|
coenzyme Q6 monooxygenase |
| chr8_+_85428391 | 0.07 |
ENSMUST00000238338.2
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr8_-_96161414 | 0.07 |
ENSMUST00000211908.2
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr1_+_52158599 | 0.07 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr5_+_35156389 | 0.07 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr4_+_130640611 | 0.07 |
ENSMUST00000156225.8
ENSMUST00000156742.8 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr11_+_98798627 | 0.07 |
ENSMUST00000092706.13
|
Cdc6
|
cell division cycle 6 |
| chr7_-_126641565 | 0.07 |
ENSMUST00000205806.2
|
Kif22
|
kinesin family member 22 |
| chr9_-_44272923 | 0.07 |
ENSMUST00000034644.10
|
Vps11
|
VPS11, CORVET/HOPS core subunit |
| chr1_+_75498162 | 0.07 |
ENSMUST00000027414.16
ENSMUST00000113553.2 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr5_+_124690908 | 0.07 |
ENSMUST00000071057.14
ENSMUST00000111438.2 |
Ddx55
|
DEAD box helicase 55 |
| chr1_-_170803680 | 0.07 |
ENSMUST00000027966.14
ENSMUST00000081103.12 ENSMUST00000159688.2 |
Fcgr2b
|
Fc receptor, IgG, low affinity IIb |
| chr1_+_155034452 | 0.07 |
ENSMUST00000027743.13
ENSMUST00000195302.6 |
Stx6
|
syntaxin 6 |
| chr14_-_47514308 | 0.07 |
ENSMUST00000111792.9
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chrX_-_135641869 | 0.07 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr3_+_103875261 | 0.07 |
ENSMUST00000117150.8
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr5_-_130031842 | 0.07 |
ENSMUST00000026613.14
|
Gusb
|
glucuronidase, beta |
| chrX_+_106132840 | 0.07 |
ENSMUST00000118666.8
ENSMUST00000053375.4 |
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr4_-_86775602 | 0.07 |
ENSMUST00000102814.5
|
Rps6
|
ribosomal protein S6 |
| chr3_+_129326285 | 0.07 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr5_+_67464284 | 0.07 |
ENSMUST00000113676.6
ENSMUST00000162372.8 |
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr9_-_117979147 | 0.07 |
ENSMUST00000215799.2
ENSMUST00000044220.11 |
Cmc1
|
COX assembly mitochondrial protein 1 |
| chr1_+_164135230 | 0.07 |
ENSMUST00000193683.6
|
Nme7
|
NME/NM23 family member 7 |
| chr12_-_79027531 | 0.07 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr11_-_79418500 | 0.07 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chr13_+_108180957 | 0.06 |
ENSMUST00000095458.6
ENSMUST00000223808.2 ENSMUST00000226042.2 ENSMUST00000225972.2 |
Smim15
|
small integral membrane protein 15 |
| chr11_-_53191208 | 0.06 |
ENSMUST00000020630.8
|
Hspa4
|
heat shock protein 4 |
| chr11_+_88095206 | 0.06 |
ENSMUST00000024486.14
|
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr1_-_167112784 | 0.06 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr4_+_118266582 | 0.06 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr15_+_25774070 | 0.06 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr9_+_89081407 | 0.06 |
ENSMUST00000138109.2
|
Gm29094
|
predicted gene 29094 |
| chr4_-_116484675 | 0.06 |
ENSMUST00000081182.5
ENSMUST00000030457.12 |
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr17_+_34149820 | 0.06 |
ENSMUST00000234226.2
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr12_-_44256843 | 0.06 |
ENSMUST00000220421.2
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chrX_-_56371953 | 0.06 |
ENSMUST00000176986.8
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr6_-_124715542 | 0.06 |
ENSMUST00000174265.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr1_-_144124871 | 0.06 |
ENSMUST00000189061.7
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr13_-_14237958 | 0.06 |
ENSMUST00000223174.2
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr19_-_45580258 | 0.06 |
ENSMUST00000160003.8
ENSMUST00000162879.8 |
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr17_+_57586094 | 0.06 |
ENSMUST00000169220.9
ENSMUST00000005889.16 ENSMUST00000112870.5 |
Vav1
|
vav 1 oncogene |
| chr5_-_121665249 | 0.06 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr2_+_181138958 | 0.06 |
ENSMUST00000149163.8
ENSMUST00000000844.15 ENSMUST00000184849.8 ENSMUST00000108800.8 ENSMUST00000069712.9 |
Tpd52l2
|
tumor protein D52-like 2 |
| chr14_+_103284413 | 0.06 |
ENSMUST00000022722.7
|
Acod1
|
aconitate decarboxylase 1 |
| chr1_-_58009191 | 0.06 |
ENSMUST00000159826.2
ENSMUST00000164963.8 |
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr3_-_53771185 | 0.06 |
ENSMUST00000122330.2
ENSMUST00000146598.8 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr2_+_118943274 | 0.06 |
ENSMUST00000140939.8
ENSMUST00000028795.10 |
Rad51
|
RAD51 recombinase |
| chr11_+_6510167 | 0.06 |
ENSMUST00000109722.9
|
Ccm2
|
cerebral cavernous malformation 2 |
| chr12_+_28801802 | 0.06 |
ENSMUST00000221877.2
ENSMUST00000222407.2 ENSMUST00000221555.2 |
Eipr1
|
EARP complex and GARP complex interacting protein 1 |
| chr10_-_89568106 | 0.06 |
ENSMUST00000020109.5
|
Actr6
|
ARP6 actin-related protein 6 |
| chr6_+_48661503 | 0.06 |
ENSMUST00000067506.14
ENSMUST00000119575.8 ENSMUST00000121957.8 ENSMUST00000090070.6 |
Gimap4
|
GTPase, IMAP family member 4 |
| chr13_+_98399693 | 0.06 |
ENSMUST00000091356.11
ENSMUST00000123924.8 |
Ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr19_-_6887361 | 0.06 |
ENSMUST00000025904.12
|
Prdx5
|
peroxiredoxin 5 |
| chr5_+_31609802 | 0.06 |
ENSMUST00000202244.4
|
Zfp512
|
zinc finger protein 512 |
| chr11_-_120348762 | 0.06 |
ENSMUST00000137632.2
ENSMUST00000044007.3 |
Oxld1
|
oxidoreductase like domain containing 1 |
| chr10_-_44334711 | 0.06 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr2_-_149978560 | 0.06 |
ENSMUST00000094538.6
ENSMUST00000109931.8 ENSMUST00000089207.13 |
Zfp120
|
zinc finger protein 120 |
| chr16_+_36755338 | 0.06 |
ENSMUST00000023531.15
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr15_-_98729333 | 0.06 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr15_+_38976260 | 0.06 |
ENSMUST00000022909.10
|
Dcaf13
|
DDB1 and CUL4 associated factor 13 |
| chr1_+_52158693 | 0.06 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chrX_+_70408351 | 0.06 |
ENSMUST00000146213.8
ENSMUST00000114601.8 ENSMUST00000015358.8 |
Mtmr1
|
myotubularin related protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spic
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.2 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.1 | 0.2 | GO:0032618 | interleukin-15 production(GO:0032618) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.1 | 0.5 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.2 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.1 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.0 | 0.2 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) negative regulation of hypersensitivity(GO:0002884) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.0 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.2 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.2 | GO:0070893 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.1 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.0 | 0.1 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0071931 | Cajal body organization(GO:0030576) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.1 | GO:0070358 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.0 | 0.1 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.0 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) |
| 0.0 | 0.0 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.0 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) protein lipidation involved in autophagosome assembly(GO:0061739) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.1 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.0 | 0.2 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.6 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.6 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |