Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
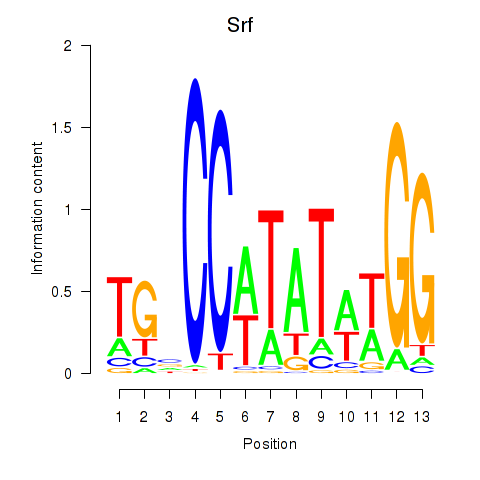
Results for Srf
Z-value: 1.22
Transcription factors associated with Srf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Srf
|
ENSMUSG00000015605.7 | serum response factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srf | mm39_v1_chr17_-_46867083_46867114 | -0.95 | 1.2e-02 | Click! |
Activity profile of Srf motif
Sorted Z-values of Srf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_34994253 | 1.75 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr13_+_23930717 | 1.19 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr11_-_73215442 | 0.99 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr7_+_3341597 | 0.71 |
ENSMUST00000164553.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr10_-_111833138 | 0.71 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr16_-_4698148 | 0.60 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr7_-_19043955 | 0.46 |
ENSMUST00000207334.2
ENSMUST00000208505.2 ENSMUST00000207716.2 ENSMUST00000208326.2 ENSMUST00000003640.4 |
Fosb
|
FBJ osteosarcoma oncogene B |
| chr2_-_92290054 | 0.45 |
ENSMUST00000136718.2
|
Slc35c1
|
solute carrier family 35, member C1 |
| chr2_-_13276074 | 0.44 |
ENSMUST00000137670.3
ENSMUST00000114791.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr2_-_13276205 | 0.42 |
ENSMUST00000191959.6
ENSMUST00000028059.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr2_+_164782642 | 0.37 |
ENSMUST00000137626.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr6_+_42326528 | 0.37 |
ENSMUST00000203329.3
|
Zyx
|
zyxin |
| chr2_+_143757193 | 0.37 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr1_+_39026887 | 0.37 |
ENSMUST00000194552.2
|
Pdcl3
|
phosducin-like 3 |
| chr10_+_67373691 | 0.34 |
ENSMUST00000048289.14
ENSMUST00000130933.2 ENSMUST00000105438.9 ENSMUST00000146986.2 |
Egr2
|
early growth response 2 |
| chr10_+_79824418 | 0.31 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr16_+_24266829 | 0.27 |
ENSMUST00000078988.10
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr7_+_119495058 | 0.27 |
ENSMUST00000106518.9
ENSMUST00000207270.2 ENSMUST00000208424.2 ENSMUST00000208202.2 ENSMUST00000054440.11 |
Lyrm1
|
LYR motif containing 1 |
| chr9_+_53448322 | 0.26 |
ENSMUST00000035850.8
|
Npat
|
nuclear protein in the AT region |
| chr11_+_98754434 | 0.26 |
ENSMUST00000142414.8
ENSMUST00000037480.9 |
Wipf2
|
WAS/WASL interacting protein family, member 2 |
| chr15_-_77726333 | 0.25 |
ENSMUST00000016771.13
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr15_-_76113692 | 0.23 |
ENSMUST00000074834.12
|
Plec
|
plectin |
| chr6_-_113354468 | 0.21 |
ENSMUST00000099118.8
|
Tada3
|
transcriptional adaptor 3 |
| chr7_+_119495515 | 0.21 |
ENSMUST00000106517.9
|
Lyrm1
|
LYR motif containing 1 |
| chr5_+_98328723 | 0.20 |
ENSMUST00000112959.4
|
Prdm8
|
PR domain containing 8 |
| chr4_+_43957401 | 0.20 |
ENSMUST00000030202.14
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr6_+_136509922 | 0.20 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr4_-_141327146 | 0.19 |
ENSMUST00000141518.8
ENSMUST00000127455.8 ENSMUST00000105784.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr4_-_122779837 | 0.18 |
ENSMUST00000106255.8
ENSMUST00000106257.10 |
Cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr11_-_120239301 | 0.18 |
ENSMUST00000062147.14
ENSMUST00000128055.2 |
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr15_-_36794741 | 0.18 |
ENSMUST00000110361.8
ENSMUST00000022894.14 ENSMUST00000110359.2 |
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr6_-_113354337 | 0.18 |
ENSMUST00000043333.9
|
Tada3
|
transcriptional adaptor 3 |
| chr6_-_113354826 | 0.17 |
ENSMUST00000032410.14
|
Tada3
|
transcriptional adaptor 3 |
| chr11_-_120622770 | 0.17 |
ENSMUST00000154565.2
ENSMUST00000026148.9 |
Cbr2
|
carbonyl reductase 2 |
| chr14_+_70314727 | 0.16 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr2_+_164675697 | 0.15 |
ENSMUST00000143780.9
|
Ctsa
|
cathepsin A |
| chr11_-_69553451 | 0.14 |
ENSMUST00000018905.12
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr13_+_109397184 | 0.14 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr4_+_101843823 | 0.14 |
ENSMUST00000106914.8
|
Gm12789
|
predicted gene 12789 |
| chr14_+_20979466 | 0.14 |
ENSMUST00000022369.9
|
Vcl
|
vinculin |
| chr7_+_44647072 | 0.14 |
ENSMUST00000003284.16
ENSMUST00000209066.2 |
Irf3
|
interferon regulatory factor 3 |
| chr19_-_5962862 | 0.14 |
ENSMUST00000136983.8
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr8_+_95113066 | 0.13 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr2_-_60503998 | 0.12 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr5_-_106844685 | 0.12 |
ENSMUST00000127434.8
ENSMUST00000112696.8 ENSMUST00000112698.8 |
Zfp644
|
zinc finger protein 644 |
| chr5_-_106844396 | 0.12 |
ENSMUST00000137285.8
ENSMUST00000124263.2 ENSMUST00000112695.4 ENSMUST00000155495.8 ENSMUST00000135108.2 ENSMUST00000149128.3 |
Zfp644
Gm28039
|
zinc finger protein 644 predicted gene, 28039 |
| chr17_+_8144822 | 0.12 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr1_+_74448535 | 0.12 |
ENSMUST00000027366.13
|
Vil1
|
villin 1 |
| chr17_+_24022153 | 0.11 |
ENSMUST00000190686.7
ENSMUST00000088621.11 ENSMUST00000233636.2 |
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr7_-_44646960 | 0.11 |
ENSMUST00000207443.2
ENSMUST00000207755.2 ENSMUST00000003290.12 |
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr15_-_99717956 | 0.10 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr4_+_137589548 | 0.10 |
ENSMUST00000102518.10
|
Ece1
|
endothelin converting enzyme 1 |
| chr6_+_113355076 | 0.10 |
ENSMUST00000156898.5
ENSMUST00000203578.3 ENSMUST00000171058.8 |
Arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr8_-_124621483 | 0.09 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chr11_-_70560110 | 0.08 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr11_-_120239339 | 0.08 |
ENSMUST00000071555.13
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr10_-_120735000 | 0.08 |
ENSMUST00000092143.12
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr11_+_82802079 | 0.08 |
ENSMUST00000018989.14
ENSMUST00000164945.3 |
Unc45b
|
unc-45 myosin chaperone B |
| chr7_+_120276801 | 0.08 |
ENSMUST00000208454.2
ENSMUST00000060175.8 |
Mosmo
|
modulator of smoothened |
| chr12_-_40249489 | 0.07 |
ENSMUST00000220951.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr6_-_71239216 | 0.07 |
ENSMUST00000129630.3
ENSMUST00000114186.9 ENSMUST00000074301.10 |
Smyd1
|
SET and MYND domain containing 1 |
| chr4_-_141327253 | 0.07 |
ENSMUST00000147785.8
|
Fblim1
|
filamin binding LIM protein 1 |
| chr6_-_86503178 | 0.07 |
ENSMUST00000053015.7
|
Pcbp1
|
poly(rC) binding protein 1 |
| chr11_-_120238917 | 0.07 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr5_+_24618380 | 0.06 |
ENSMUST00000049346.10
|
Asic3
|
acid-sensing (proton-gated) ion channel 3 |
| chr14_-_19420488 | 0.06 |
ENSMUST00000166494.2
|
Gm2897
|
predicted gene 2897 |
| chr14_+_16182428 | 0.06 |
ENSMUST00000170104.3
|
Gm3411
|
predicted gene 3411 |
| chr6_-_36787096 | 0.05 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr14_-_17398733 | 0.05 |
ENSMUST00000163719.8
|
Gm8281
|
predicted gene, 8281 |
| chr1_-_44118902 | 0.05 |
ENSMUST00000238662.2
|
Gm8251
|
predicted gene 8251 |
| chr14_+_53315909 | 0.05 |
ENSMUST00000103608.4
|
Trav14d-3-dv8
|
T cell receptor alpha variable 14D-3-DV8 |
| chr7_-_30062197 | 0.05 |
ENSMUST00000046351.7
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr13_+_52000704 | 0.04 |
ENSMUST00000021903.3
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr2_-_113883285 | 0.04 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr14_-_34310602 | 0.04 |
ENSMUST00000064098.14
ENSMUST00000090040.12 ENSMUST00000022330.9 ENSMUST00000022327.13 |
Ldb3
|
LIM domain binding 3 |
| chr11_+_77353431 | 0.04 |
ENSMUST00000130255.2
|
Coro6
|
coronin 6 |
| chr14_-_18817743 | 0.04 |
ENSMUST00000167430.8
|
Gm3020
|
predicted gene 3020 |
| chr4_-_119047167 | 0.04 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr11_+_33913013 | 0.04 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr14_-_19057159 | 0.04 |
ENSMUST00000170123.2
|
Gm10409
|
predicted gene 10409 |
| chr6_-_85490568 | 0.04 |
ENSMUST00000095759.5
|
Egr4
|
early growth response 4 |
| chr19_+_5497575 | 0.04 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr19_+_12394797 | 0.04 |
ENSMUST00000216506.2
ENSMUST00000214153.2 |
Olfr1441
|
olfactory receptor 1441 |
| chr14_+_14901127 | 0.04 |
ENSMUST00000163790.2
|
Gm3558
|
predicted gene 3558 |
| chr4_+_143785696 | 0.03 |
ENSMUST00000091676.12
ENSMUST00000133626.8 |
Pramel4
|
PRAME like 4 |
| chr14_-_19261196 | 0.03 |
ENSMUST00000112797.12
ENSMUST00000225885.2 |
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chr14_-_18359247 | 0.03 |
ENSMUST00000170207.8
|
Gm8108
|
predicted gene 8108 |
| chr7_-_101859379 | 0.03 |
ENSMUST00000210682.2
|
Nup98
|
nucleoporin 98 |
| chr14_-_18897750 | 0.03 |
ENSMUST00000178728.2
|
Gm3005
|
predicted gene 3005 |
| chr17_-_73706284 | 0.03 |
ENSMUST00000095208.4
|
Capn13
|
calpain 13 |
| chr14_-_18287197 | 0.03 |
ENSMUST00000164512.8
|
Gm2974
|
predicted gene 2974 |
| chr14_-_16968099 | 0.03 |
ENSMUST00000181562.8
|
Gm3488
|
predicted gene, 3488 |
| chr7_+_17743802 | 0.03 |
ENSMUST00000081703.8
ENSMUST00000108488.7 |
Ceacam13
|
carcinoembryonic antigen-related cell adhesion molecule 13 |
| chr14_-_55204092 | 0.03 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr11_+_117157024 | 0.03 |
ENSMUST00000019038.15
|
Septin9
|
septin 9 |
| chr12_+_108389075 | 0.03 |
ENSMUST00000109860.8
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr7_-_101859308 | 0.03 |
ENSMUST00000070165.7
ENSMUST00000211235.2 ENSMUST00000211022.2 |
Nup98
|
nucleoporin 98 |
| chr14_+_70314652 | 0.03 |
ENSMUST00000035908.3
|
Egr3
|
early growth response 3 |
| chr16_+_15455707 | 0.03 |
ENSMUST00000023352.9
|
Prkdc
|
protein kinase, DNA activated, catalytic polypeptide |
| chr1_-_127605660 | 0.03 |
ENSMUST00000160616.8
|
Tmem163
|
transmembrane protein 163 |
| chr14_+_33662976 | 0.03 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr7_-_142215027 | 0.03 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr10_+_45211847 | 0.02 |
ENSMUST00000095715.5
|
Bves
|
blood vessel epicardial substance |
| chr7_+_17706049 | 0.02 |
ENSMUST00000094799.3
|
Ceacam11
|
carcinoembryonic antigen-related cell adhesion molecule 11 |
| chr14_-_19635203 | 0.02 |
ENSMUST00000170694.9
|
Gm2237
|
predicted gene 2237 |
| chr14_-_17742998 | 0.02 |
ENSMUST00000165619.8
|
Gm3252
|
predicted gene 3252 |
| chr13_-_21823691 | 0.02 |
ENSMUST00000043081.3
|
Olfr11
|
olfactory receptor 11 |
| chr14_-_55204054 | 0.02 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr9_-_58220469 | 0.02 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr14_+_15295240 | 0.02 |
ENSMUST00000172431.8
|
Gm3512
|
predicted gene 3512 |
| chrY_-_10147183 | 0.02 |
ENSMUST00000189630.2
|
Gm21310
|
predicted gene, 21310 |
| chr14_+_16508028 | 0.02 |
ENSMUST00000163885.2
|
Gm3248
|
predicted gene 3248 |
| chr14_-_19137146 | 0.02 |
ENSMUST00000177786.8
|
Gm2956
|
predicted gene 2956 |
| chr14_+_16589391 | 0.02 |
ENSMUST00000164484.8
|
Gm8237
|
predicted gene 8237 |
| chrX_+_85235370 | 0.02 |
ENSMUST00000026036.5
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr9_+_110592709 | 0.02 |
ENSMUST00000079784.12
|
Myl3
|
myosin, light polypeptide 3 |
| chr14_-_18659699 | 0.02 |
ENSMUST00000170480.8
|
Gm3002
|
predicted gene 3002 |
| chr14_-_18054325 | 0.02 |
ENSMUST00000168866.2
|
Gm3164
|
predicted gene 3164 |
| chr14_+_15579811 | 0.02 |
ENSMUST00000171906.2
|
Gm3667
|
predicted gene 3667 |
| chr2_+_89842475 | 0.02 |
ENSMUST00000214382.2
ENSMUST00000217065.3 |
Olfr1263
|
olfactory receptor 1263 |
| chr6_-_97156032 | 0.02 |
ENSMUST00000095664.6
|
Tmf1
|
TATA element modulatory factor 1 |
| chr6_-_132187484 | 0.02 |
ENSMUST00000087853.4
|
Prb1
|
proline-rich protein BstNI subfamily 1 |
| chr7_-_20208850 | 0.02 |
ENSMUST00000098744.2
|
Gm10670
|
predicted gene 10670 |
| chr11_-_12362136 | 0.02 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr7_-_119495508 | 0.02 |
ENSMUST00000106519.8
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr7_-_20832438 | 0.02 |
ENSMUST00000105197.3
|
Vmn1r121
|
vomeronasal 1 receptor 121 |
| chr11_+_101221431 | 0.01 |
ENSMUST00000103105.10
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr2_-_103627937 | 0.01 |
ENSMUST00000028607.13
|
Caprin1
|
cell cycle associated protein 1 |
| chrX_-_56549092 | 0.01 |
ENSMUST00000057645.6
|
Gpr101
|
G protein-coupled receptor 101 |
| chr14_+_15442324 | 0.01 |
ENSMUST00000170738.3
|
Gm10406
|
predicted gene 10406 |
| chr14_-_17614197 | 0.01 |
ENSMUST00000166776.8
|
Gm3264
|
predicted gene 3264 |
| chr1_+_167445815 | 0.01 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr18_+_6603627 | 0.01 |
ENSMUST00000234821.2
ENSMUST00000044829.5 ENSMUST00000234337.2 |
4921524L21Rik
|
RIKEN cDNA 4921524L21 gene |
| chr9_-_78263025 | 0.01 |
ENSMUST00000125479.8
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr14_-_18135010 | 0.01 |
ENSMUST00000165466.8
|
2610042L04Rik
|
RIKEN cDNA 2610042L04 gene |
| chr7_-_21035392 | 0.01 |
ENSMUST00000168398.2
|
Vmn1r126
|
vomeronasal 1 receptor 126 |
| chr14_+_16728196 | 0.01 |
ENSMUST00000177556.8
|
Gm3373
|
predicted gene 3373 |
| chr4_+_39450265 | 0.01 |
ENSMUST00000029955.5
|
1700009N14Rik
|
RIKEN cDNA 1700009N14 gene |
| chr17_-_37508902 | 0.01 |
ENSMUST00000055324.8
|
Olfr94
|
olfactory receptor 94 |
| chr14_+_17080724 | 0.01 |
ENSMUST00000177986.8
|
Gm3500
|
predicted gene 3500 |
| chr14_+_15154724 | 0.01 |
ENSMUST00000165744.2
|
Gm3739
|
predicted gene 3739 |
| chr14_+_16361108 | 0.01 |
ENSMUST00000165193.2
|
Gm3468
|
predicted gene 3468 |
| chrX_-_165301563 | 0.01 |
ENSMUST00000112187.2
|
Tceanc
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr14_-_34224479 | 0.01 |
ENSMUST00000171551.2
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
| chr14_-_34310438 | 0.01 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr15_+_11000802 | 0.01 |
ENSMUST00000117100.4
|
Slc45a2
|
solute carrier family 45, member 2 |
| chr18_+_45402018 | 0.01 |
ENSMUST00000183850.8
ENSMUST00000066890.14 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr14_-_34310637 | 0.01 |
ENSMUST00000227819.2
|
Ldb3
|
LIM domain binding 3 |
| chr14_-_45767421 | 0.01 |
ENSMUST00000150660.3
|
Fermt2
|
fermitin family member 2 |
| chr6_-_83513222 | 0.01 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr7_-_105249308 | 0.01 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr7_+_15863679 | 0.01 |
ENSMUST00000211649.2
|
Slc8a2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr14_+_16431564 | 0.01 |
ENSMUST00000164139.2
|
Gm8206
|
predicted gene 8206 |
| chr6_-_83513184 | 0.01 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr3_-_93846078 | 0.01 |
ENSMUST00000177735.2
|
Tdpoz9-ps1
|
TD and POZ domain containing 9, pseudogene 1 |
| chr16_-_10614679 | 0.01 |
ENSMUST00000023144.6
|
Prm1
|
protamine 1 |
| chr16_-_58747023 | 0.01 |
ENSMUST00000205742.2
|
Olfr181
|
olfactory receptor 181 |
| chrX_+_11178173 | 0.01 |
ENSMUST00000178979.2
|
H2al1e
|
H2A histone family member L1E |
| chr9_+_57913694 | 0.00 |
ENSMUST00000188116.7
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr17_+_37756371 | 0.00 |
ENSMUST00000078207.4
ENSMUST00000218675.2 |
Olfr108
|
olfactory receptor 108 |
| chr7_-_22543030 | 0.00 |
ENSMUST00000167871.2
|
Vmn1r159
|
vomeronasal 1 receptor 159 |
| chr7_-_142215595 | 0.00 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr4_-_96029375 | 0.00 |
ENSMUST00000097972.5
|
Cyp2j12
|
cytochrome P450, family 2, subfamily j, polypeptide 12 |
| chr5_-_24534554 | 0.00 |
ENSMUST00000115098.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr9_+_74959259 | 0.00 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr16_-_92262969 | 0.00 |
ENSMUST00000232239.2
ENSMUST00000060005.15 |
Rcan1
|
regulator of calcineurin 1 |
| chr17_+_38646050 | 0.00 |
ENSMUST00000077203.3
|
Olfr136
|
olfactory receptor 136 |
| chr8_+_55003359 | 0.00 |
ENSMUST00000033918.4
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr6_-_23248361 | 0.00 |
ENSMUST00000031709.7
|
Fezf1
|
Fez family zinc finger 1 |
| chr5_+_88117307 | 0.00 |
ENSMUST00000007601.4
ENSMUST00000187738.2 |
2310003L06Rik
Gm28434
|
RIKEN cDNA 2310003L06 gene predicted gene 28434 |
| chr15_-_77129786 | 0.00 |
ENSMUST00000228558.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_-_5962798 | 0.00 |
ENSMUST00000118623.2
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr12_-_40249314 | 0.00 |
ENSMUST00000095760.3
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr15_-_77129706 | 0.00 |
ENSMUST00000228361.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr10_+_129603367 | 0.00 |
ENSMUST00000060636.2
|
Olfr808
|
olfactory receptor 808 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Srf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.1 | 1.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.3 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.1 | 0.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.4 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.0 | 0.1 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.0 | 0.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.0 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0021957 | corpus callosum morphogenesis(GO:0021540) corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.3 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.3 | 1.8 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |