Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
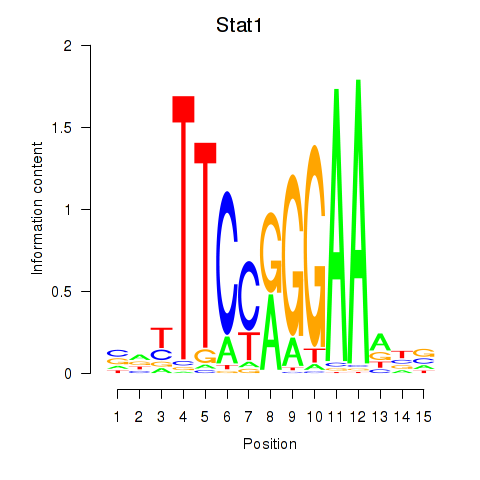
Results for Stat1
Z-value: 1.83
Transcription factors associated with Stat1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat1
|
ENSMUSG00000026104.15 | signal transducer and activator of transcription 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat1 | mm39_v1_chr1_+_52158721_52158783 | 1.00 | 8.4e-05 | Click! |
Activity profile of Stat1 motif
Sorted Z-values of Stat1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_142202642 | 1.92 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr7_+_43086432 | 1.69 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr9_+_110867807 | 1.43 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr17_+_29309942 | 1.28 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr17_-_34406193 | 1.21 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr17_-_56428968 | 1.17 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr7_+_43086554 | 1.16 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr12_-_108859123 | 1.10 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr10_-_17898938 | 1.01 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chr1_+_134110142 | 0.95 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr2_-_65955338 | 0.83 |
ENSMUST00000028378.4
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr2_-_163592127 | 0.79 |
ENSMUST00000017841.4
|
Ada
|
adenosine deaminase |
| chr12_-_112637998 | 0.77 |
ENSMUST00000128300.9
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr4_-_136613498 | 0.77 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr17_+_34406762 | 0.76 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr19_-_46950355 | 0.73 |
ENSMUST00000236501.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr17_+_34406523 | 0.71 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr10_-_79911245 | 0.66 |
ENSMUST00000217972.2
|
Sbno2
|
strawberry notch 2 |
| chr11_+_53660834 | 0.63 |
ENSMUST00000108920.10
ENSMUST00000140866.9 ENSMUST00000108922.9 |
Irf1
|
interferon regulatory factor 1 |
| chr4_+_11704438 | 0.58 |
ENSMUST00000108304.9
|
Gem
|
GTP binding protein (gene overexpressed in skeletal muscle) |
| chr15_+_34238174 | 0.58 |
ENSMUST00000022867.5
ENSMUST00000226627.2 |
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr2_+_128942900 | 0.57 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr7_+_29607917 | 0.57 |
ENSMUST00000186475.2
|
Zfp383
|
zinc finger protein 383 |
| chr8_-_112064783 | 0.56 |
ENSMUST00000056157.14
ENSMUST00000120432.3 |
Mlkl
|
mixed lineage kinase domain-like |
| chr2_+_128942919 | 0.56 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr14_+_55842002 | 0.56 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr17_-_45903410 | 0.56 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chrX_-_139857424 | 0.55 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr10_-_17898838 | 0.53 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr6_+_127423779 | 0.53 |
ENSMUST00000112191.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr8_-_112064393 | 0.52 |
ENSMUST00000145862.3
|
Mlkl
|
mixed lineage kinase domain-like |
| chr18_-_43610829 | 0.51 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr8_+_111760521 | 0.51 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr7_-_100232276 | 0.49 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr11_+_69886652 | 0.48 |
ENSMUST00000101526.9
|
Phf23
|
PHD finger protein 23 |
| chr7_+_27290969 | 0.47 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr4_+_99603877 | 0.47 |
ENSMUST00000097961.9
ENSMUST00000107004.9 ENSMUST00000139799.3 |
Alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr15_+_88960327 | 0.46 |
ENSMUST00000165690.2
|
Trabd
|
TraB domain containing |
| chr7_-_12787611 | 0.46 |
ENSMUST00000182490.2
|
Mzf1
|
myeloid zinc finger 1 |
| chr1_+_134109888 | 0.45 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr2_+_101716577 | 0.45 |
ENSMUST00000028584.8
|
Commd9
|
COMM domain containing 9 |
| chr9_-_57460004 | 0.42 |
ENSMUST00000034856.15
|
Mpi
|
mannose phosphate isomerase |
| chr3_-_96201248 | 0.40 |
ENSMUST00000029748.8
|
Fcgr1
|
Fc receptor, IgG, high affinity I |
| chr17_+_28910302 | 0.39 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr7_+_45204317 | 0.39 |
ENSMUST00000107752.12
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr7_+_29515485 | 0.39 |
ENSMUST00000178162.3
ENSMUST00000032796.14 |
Zfp790
|
zinc finger protein 790 |
| chr7_-_99770653 | 0.38 |
ENSMUST00000208670.2
ENSMUST00000032969.14 |
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr2_+_150590956 | 0.38 |
ENSMUST00000094467.6
|
Entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 |
| chr6_-_48025845 | 0.38 |
ENSMUST00000095944.10
|
Zfp777
|
zinc finger protein 777 |
| chr2_-_126341757 | 0.38 |
ENSMUST00000040128.12
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr7_+_27291126 | 0.37 |
ENSMUST00000167435.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr19_+_36811615 | 0.36 |
ENSMUST00000025729.12
|
Tnks2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr10_-_17898977 | 0.36 |
ENSMUST00000020002.9
|
Abracl
|
ABRA C-terminal like |
| chrX_+_105059305 | 0.36 |
ENSMUST00000033582.5
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr16_-_35691914 | 0.35 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr19_+_37184927 | 0.35 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr5_+_52991351 | 0.35 |
ENSMUST00000031072.14
|
Anapc4
|
anaphase promoting complex subunit 4 |
| chr11_-_75081284 | 0.35 |
ENSMUST00000044949.11
|
Dph1
|
diphthamide biosynthesis 1 |
| chr4_+_63478478 | 0.34 |
ENSMUST00000080336.4
|
Tmem268
|
transmembrane protein 268 |
| chr7_-_108774367 | 0.34 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr14_+_56905698 | 0.34 |
ENSMUST00000116468.2
|
Mphosph8
|
M-phase phosphoprotein 8 |
| chr3_-_144412394 | 0.34 |
ENSMUST00000200532.2
|
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr16_-_91728162 | 0.34 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr4_-_129534403 | 0.33 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr9_-_107158058 | 0.33 |
ENSMUST00000134682.3
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr7_-_24016020 | 0.32 |
ENSMUST00000108436.8
ENSMUST00000032673.15 |
Zfp94
|
zinc finger protein 94 |
| chr16_+_17326810 | 0.32 |
ENSMUST00000231292.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr11_-_72106418 | 0.32 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr8_+_106052970 | 0.31 |
ENSMUST00000015000.12
ENSMUST00000098453.9 |
Tmem208
|
transmembrane protein 208 |
| chr8_+_121463090 | 0.31 |
ENSMUST00000160943.3
ENSMUST00000047737.10 ENSMUST00000162658.8 |
Irf8
|
interferon regulatory factor 8 |
| chr16_+_19578981 | 0.30 |
ENSMUST00000079780.10
ENSMUST00000164397.8 |
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr5_-_5564730 | 0.30 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr1_+_171041539 | 0.29 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr2_-_127426588 | 0.29 |
ENSMUST00000110368.9
ENSMUST00000077422.12 |
Zfp661
|
zinc finger protein 661 |
| chr3_+_84832783 | 0.28 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr7_+_29794575 | 0.28 |
ENSMUST00000130526.2
ENSMUST00000108200.2 |
Zfp260
|
zinc finger protein 260 |
| chr7_-_12788441 | 0.27 |
ENSMUST00000182087.2
|
Mzf1
|
myeloid zinc finger 1 |
| chr17_-_45903494 | 0.27 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr2_-_84605732 | 0.26 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chrX_+_158086253 | 0.26 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr16_+_17327076 | 0.26 |
ENSMUST00000232242.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr11_-_59937302 | 0.26 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr11_+_81926394 | 0.25 |
ENSMUST00000000193.6
|
Ccl2
|
chemokine (C-C motif) ligand 2 |
| chr1_+_84936703 | 0.25 |
ENSMUST00000239195.2
|
Gm15433
|
predicted pseudogene 15433 |
| chr17_+_28910393 | 0.24 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr7_+_79836581 | 0.23 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr18_+_21135079 | 0.23 |
ENSMUST00000234545.2
|
Rnf138
|
ring finger protein 138 |
| chr4_-_129534853 | 0.22 |
ENSMUST00000046425.16
ENSMUST00000133803.8 |
Txlna
|
taxilin alpha |
| chr7_-_99770280 | 0.22 |
ENSMUST00000208184.2
|
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr13_-_85437118 | 0.22 |
ENSMUST00000109552.3
|
Rasa1
|
RAS p21 protein activator 1 |
| chr7_-_81479179 | 0.22 |
ENSMUST00000026093.9
|
Btbd1
|
BTB (POZ) domain containing 1 |
| chr7_+_65343156 | 0.21 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr13_-_85437201 | 0.21 |
ENSMUST00000223598.2
|
Rasa1
|
RAS p21 protein activator 1 |
| chr2_-_155571279 | 0.20 |
ENSMUST00000040833.5
|
Edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr3_-_51184730 | 0.20 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr8_+_11777721 | 0.19 |
ENSMUST00000210104.2
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr4_-_129534752 | 0.19 |
ENSMUST00000132217.8
ENSMUST00000130017.2 ENSMUST00000154105.8 |
Txlna
|
taxilin alpha |
| chr12_-_11315755 | 0.19 |
ENSMUST00000166117.4
ENSMUST00000219600.2 ENSMUST00000218487.2 |
Gen1
|
GEN1, Holliday junction 5' flap endonuclease |
| chr8_-_111808488 | 0.19 |
ENSMUST00000188466.7
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr8_-_111808674 | 0.19 |
ENSMUST00000039597.14
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr9_-_59393893 | 0.18 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chrX_-_72974357 | 0.17 |
ENSMUST00000155597.2
ENSMUST00000114379.8 |
Renbp
|
renin binding protein |
| chr4_+_120523758 | 0.17 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr16_+_19578945 | 0.17 |
ENSMUST00000121344.8
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr11_+_101315893 | 0.17 |
ENSMUST00000040561.6
|
Rundc1
|
RUN domain containing 1 |
| chr3_-_90398270 | 0.17 |
ENSMUST00000149884.2
|
Snapin
|
SNAP-associated protein |
| chr16_-_17711950 | 0.16 |
ENSMUST00000155943.9
|
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr10_+_128139227 | 0.16 |
ENSMUST00000218315.2
ENSMUST00000219721.2 |
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr3_-_10505113 | 0.16 |
ENSMUST00000029047.12
ENSMUST00000195822.2 ENSMUST00000099223.11 |
Snx16
|
sorting nexin 16 |
| chr13_-_67903312 | 0.16 |
ENSMUST00000144183.3
|
Zfp85
|
zinc finger protein 85 |
| chr11_-_98620200 | 0.15 |
ENSMUST00000126565.2
ENSMUST00000100500.9 ENSMUST00000017354.13 |
Med24
|
mediator complex subunit 24 |
| chr2_+_3425159 | 0.15 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr3_-_37180093 | 0.15 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr2_-_84605764 | 0.15 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr7_+_23969822 | 0.15 |
ENSMUST00000108438.10
|
Zfp93
|
zinc finger protein 93 |
| chr12_+_108859557 | 0.14 |
ENSMUST00000221377.2
|
Wdr25
|
WD repeat domain 25 |
| chr5_+_134212836 | 0.14 |
ENSMUST00000016086.10
|
Gtf2ird2
|
GTF2I repeat domain containing 2 |
| chrX_-_8011952 | 0.14 |
ENSMUST00000115615.9
ENSMUST00000115616.8 ENSMUST00000115621.9 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr4_+_63478454 | 0.14 |
ENSMUST00000124332.8
ENSMUST00000150360.8 |
Tmem268
|
transmembrane protein 268 |
| chr3_-_51184895 | 0.14 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr10_+_128139191 | 0.14 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr10_+_126877798 | 0.13 |
ENSMUST00000006915.14
ENSMUST00000120542.8 |
Mettl1
|
methyltransferase like 1 |
| chr2_-_120801186 | 0.13 |
ENSMUST00000028728.6
|
Ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chrX_-_72974440 | 0.13 |
ENSMUST00000116578.8
|
Renbp
|
renin binding protein |
| chr11_-_101315345 | 0.12 |
ENSMUST00000107257.8
ENSMUST00000107259.4 ENSMUST00000107252.9 ENSMUST00000093933.11 |
Gm27029
Ptges3l
|
predicted gene, 27029 prostaglandin E synthase 3 like |
| chr16_-_31898088 | 0.12 |
ENSMUST00000023467.9
|
Pak2
|
p21 (RAC1) activated kinase 2 |
| chr5_+_14075281 | 0.12 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr3_+_122039274 | 0.12 |
ENSMUST00000178826.8
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr12_+_86129329 | 0.12 |
ENSMUST00000054565.8
ENSMUST00000222821.2 ENSMUST00000222905.2 |
Ift43
|
intraflagellar transport 43 |
| chr16_-_20245138 | 0.11 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr4_-_63965161 | 0.11 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr3_+_99161070 | 0.11 |
ENSMUST00000029462.10
|
Tbx15
|
T-box 15 |
| chr9_+_38629560 | 0.10 |
ENSMUST00000001544.12
ENSMUST00000118144.8 |
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr11_+_81992662 | 0.10 |
ENSMUST00000000194.4
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr15_+_25933632 | 0.10 |
ENSMUST00000228327.2
|
Retreg1
|
reticulophagy regulator 1 |
| chrX_-_8118541 | 0.09 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr4_+_148686985 | 0.09 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr11_+_69886603 | 0.09 |
ENSMUST00000018716.10
|
Phf23
|
PHD finger protein 23 |
| chr7_+_45204350 | 0.09 |
ENSMUST00000210300.2
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr6_+_34575435 | 0.09 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr9_+_107173907 | 0.08 |
ENSMUST00000168260.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr6_-_65121892 | 0.08 |
ENSMUST00000031982.5
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr4_+_114916703 | 0.08 |
ENSMUST00000162489.2
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr12_+_11315868 | 0.07 |
ENSMUST00000020931.6
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr5_-_5564873 | 0.07 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr9_+_123822000 | 0.07 |
ENSMUST00000039171.9
|
Ccr3
|
chemokine (C-C motif) receptor 3 |
| chr5_+_75312939 | 0.06 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr9_+_107174081 | 0.06 |
ENSMUST00000167072.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr18_-_3280999 | 0.06 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr8_+_71126012 | 0.06 |
ENSMUST00000146972.3
ENSMUST00000210071.2 ENSMUST00000210987.2 |
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr11_-_99877423 | 0.06 |
ENSMUST00000105050.4
|
Krtap16-1
|
keratin associated protein 16-1 |
| chr8_-_71060911 | 0.05 |
ENSMUST00000210580.2
ENSMUST00000211608.2 ENSMUST00000049908.11 |
Ssbp4
|
single stranded DNA binding protein 4 |
| chr8_-_68363564 | 0.05 |
ENSMUST00000093468.12
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_-_86640362 | 0.05 |
ENSMUST00000216117.2
|
Olfr141
|
olfactory receptor 141 |
| chr9_+_105520154 | 0.05 |
ENSMUST00000190358.2
ENSMUST00000191268.7 ENSMUST00000065778.13 ENSMUST00000188784.2 |
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr18_-_52662728 | 0.05 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
| chr5_+_90708962 | 0.05 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr2_-_101459274 | 0.05 |
ENSMUST00000099682.9
|
Iftap
|
intraflagellar transport associated protein |
| chr7_-_100505486 | 0.05 |
ENSMUST00000139604.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr7_+_91321500 | 0.04 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr18_-_3281089 | 0.04 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr5_-_34671236 | 0.04 |
ENSMUST00000114359.2
ENSMUST00000030991.14 ENSMUST00000087737.10 |
Tnip2
|
TNFAIP3 interacting protein 2 |
| chr8_-_71229293 | 0.04 |
ENSMUST00000034296.15
|
Pik3r2
|
phosphoinositide-3-kinase regulatory subunit 2 |
| chr18_-_52662917 | 0.04 |
ENSMUST00000171470.8
|
Lox
|
lysyl oxidase |
| chr4_+_114945905 | 0.03 |
ENSMUST00000171877.8
ENSMUST00000177647.8 ENSMUST00000106548.9 ENSMUST00000030488.3 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr6_-_131607875 | 0.03 |
ENSMUST00000067597.5
|
Tas2r130
|
taste receptor, type 2, member 130 |
| chr3_-_88417251 | 0.03 |
ENSMUST00000149068.2
|
Lmna
|
lamin A |
| chr13_-_42001075 | 0.03 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr17_+_36134450 | 0.03 |
ENSMUST00000172846.2
|
Flot1
|
flotillin 1 |
| chr14_-_70864666 | 0.03 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr1_-_37929414 | 0.03 |
ENSMUST00000139725.8
ENSMUST00000027257.10 |
Mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr7_-_30643444 | 0.03 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr5_+_67417908 | 0.03 |
ENSMUST00000037918.12
ENSMUST00000162543.8 ENSMUST00000161233.8 ENSMUST00000160352.8 |
Tmem33
|
transmembrane protein 33 |
| chr5_-_72893941 | 0.03 |
ENSMUST00000169534.6
|
Txk
|
TXK tyrosine kinase |
| chr2_+_87082184 | 0.03 |
ENSMUST00000081986.5
|
Olfr1115
|
olfactory receptor 1115 |
| chr2_+_177834868 | 0.03 |
ENSMUST00000103065.2
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr15_-_37458768 | 0.02 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr5_-_6926523 | 0.02 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr15_-_93417380 | 0.02 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr4_+_43401232 | 0.02 |
ENSMUST00000125399.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr10_+_129186434 | 0.02 |
ENSMUST00000077024.2
|
Olfr782
|
olfactory receptor 782 |
| chr10_-_129574784 | 0.02 |
ENSMUST00000095245.2
|
Olfr806
|
olfactory receptor 806 |
| chr6_+_21949986 | 0.02 |
ENSMUST00000149728.7
|
Ing3
|
inhibitor of growth family, member 3 |
| chr7_-_109330915 | 0.02 |
ENSMUST00000035372.3
|
Ascl3
|
achaete-scute family bHLH transcription factor 3 |
| chrX_+_7594670 | 0.02 |
ENSMUST00000033489.8
|
Praf2
|
PRA1 domain family 2 |
| chr11_-_101316156 | 0.02 |
ENSMUST00000103102.10
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr7_+_89780785 | 0.02 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr7_-_81356557 | 0.01 |
ENSMUST00000207983.2
|
Homer2
|
homer scaffolding protein 2 |
| chr11_-_58346806 | 0.01 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr7_-_81356653 | 0.01 |
ENSMUST00000026922.15
|
Homer2
|
homer scaffolding protein 2 |
| chr8_-_125296435 | 0.01 |
ENSMUST00000238882.2
ENSMUST00000063278.7 |
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr3_+_122039206 | 0.01 |
ENSMUST00000029769.14
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr10_+_69761784 | 0.01 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr9_+_7464139 | 0.01 |
ENSMUST00000217651.3
|
Mmp1a
|
matrix metallopeptidase 1a (interstitial collagenase) |
| chr18_+_52958382 | 0.01 |
ENSMUST00000238707.2
|
Gm50457
|
predicted gene, 50457 |
| chr6_+_40941688 | 0.01 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr10_+_129298547 | 0.01 |
ENSMUST00000077836.3
|
Olfr787
|
olfactory receptor 787 |
| chr1_+_36346804 | 0.01 |
ENSMUST00000126413.8
|
Arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr7_+_91321694 | 0.01 |
ENSMUST00000238608.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr10_+_129692389 | 0.01 |
ENSMUST00000064893.3
|
Olfr813
|
olfactory receptor 813 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.4 | 1.4 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.3 | 1.1 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.3 | 1.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 0.8 | GO:0046100 | germinal center B cell differentiation(GO:0002314) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 0.2 | 0.8 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 1.9 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.2 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.7 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.8 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.8 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.4 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 0.4 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 | 0.6 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 0.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.3 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 1.3 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 0.3 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) monocyte homeostasis(GO:0035702) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.1 | 0.6 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.5 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:1904154 | trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.6 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 1.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.6 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 1.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 1.1 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.4 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.3 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.5 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.3 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.0 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 1.4 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 1.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.4 | 1.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 1.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.5 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 1.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.5 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 1.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 1.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.8 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.5 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | PID IFNG PATHWAY | IFN-gamma pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 3.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |