Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
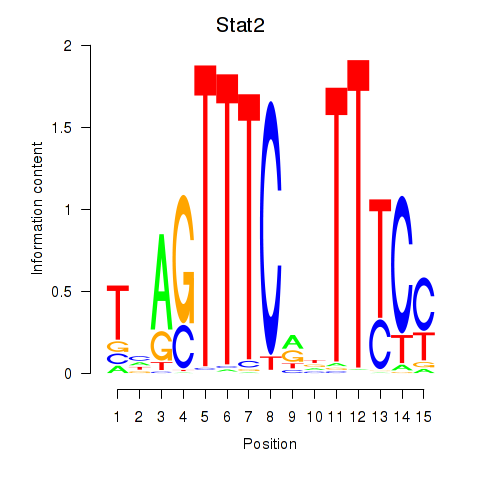
Results for Stat2
Z-value: 2.47
Transcription factors associated with Stat2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat2
|
ENSMUSG00000040033.17 | signal transducer and activator of transcription 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat2 | mm39_v1_chr10_+_128106414_128106446 | 0.98 | 3.2e-03 | Click! |
Activity profile of Stat2 motif
Sorted Z-values of Stat2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_103400470 | 3.29 |
ENSMUST00000079294.12
ENSMUST00000076788.12 ENSMUST00000076702.12 ENSMUST00000066701.13 ENSMUST00000085065.12 ENSMUST00000140838.2 |
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr8_-_112064783 | 2.46 |
ENSMUST00000056157.14
ENSMUST00000120432.3 |
Mlkl
|
mixed lineage kinase domain-like |
| chr5_-_120887864 | 1.65 |
ENSMUST00000053909.13
ENSMUST00000081491.13 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr6_+_121222865 | 1.42 |
ENSMUST00000032198.11
|
Usp18
|
ubiquitin specific peptidase 18 |
| chr17_-_34406193 | 1.33 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr11_-_118292678 | 1.32 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr5_-_121045568 | 1.26 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr8_-_112064393 | 1.16 |
ENSMUST00000145862.3
|
Mlkl
|
mixed lineage kinase domain-like |
| chr16_+_23428650 | 1.12 |
ENSMUST00000038423.6
ENSMUST00000211349.2 |
Rtp4
|
receptor transporter protein 4 |
| chr12_-_80307110 | 1.12 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr7_-_104157006 | 1.10 |
ENSMUST00000033211.14
ENSMUST00000071069.13 |
Trim30d
|
tripartite motif-containing 30D |
| chr11_-_48883053 | 1.09 |
ENSMUST00000059930.3
ENSMUST00000068063.4 |
Gm12185
Tgtp1
|
predicted gene 12185 T cell specific GTPase 1 |
| chr6_+_127423779 | 1.06 |
ENSMUST00000112191.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr1_+_85454323 | 1.01 |
ENSMUST00000239236.2
|
Gm7592
|
predicted gene 7592 |
| chr9_+_107852733 | 1.00 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr4_-_40239778 | 0.97 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr5_-_121025645 | 0.94 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr11_-_48762170 | 0.94 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr17_+_34406762 | 0.93 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr12_-_26506422 | 0.92 |
ENSMUST00000020970.10
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr16_+_26400454 | 0.92 |
ENSMUST00000096129.9
ENSMUST00000166294.9 ENSMUST00000174202.8 ENSMUST00000023156.13 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr4_-_136626073 | 0.91 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr3_+_142265787 | 0.88 |
ENSMUST00000199325.5
ENSMUST00000106222.9 |
Gbp3
|
guanylate binding protein 3 |
| chr11_+_119283887 | 0.87 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr1_-_173770010 | 0.87 |
ENSMUST00000042228.15
ENSMUST00000081216.12 ENSMUST00000129829.8 ENSMUST00000123708.8 |
Ifi203
|
interferon activated gene 203 |
| chr17_+_34406523 | 0.87 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr2_-_155668567 | 0.84 |
ENSMUST00000109638.2
ENSMUST00000134278.2 |
Eif6
|
eukaryotic translation initiation factor 6 |
| chr5_-_92496730 | 0.84 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr16_+_35759346 | 0.83 |
ENSMUST00000023622.13
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr1_+_175459559 | 0.83 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr5_+_115034997 | 0.82 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr15_-_76127600 | 0.81 |
ENSMUST00000165738.8
ENSMUST00000075689.7 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr7_+_138828391 | 0.81 |
ENSMUST00000093993.5
ENSMUST00000172136.9 |
Pwwp2b
|
PWWP domain containing 2B |
| chr19_+_3372296 | 0.80 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr2_-_105229653 | 0.79 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr16_+_35758836 | 0.79 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_+_27371168 | 0.78 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr7_+_103893658 | 0.78 |
ENSMUST00000106849.9
ENSMUST00000060315.12 |
Trim34a
|
tripartite motif-containing 34A |
| chr3_+_142265904 | 0.78 |
ENSMUST00000128609.8
ENSMUST00000029935.14 |
Gbp3
|
guanylate binding protein 3 |
| chr9_+_20779924 | 0.77 |
ENSMUST00000043911.8
|
Shfl
|
shiftless antiviral inhibitor of ribosomal frameshifting |
| chr6_+_57557978 | 0.76 |
ENSMUST00000031817.10
|
Herc6
|
hect domain and RLD 6 |
| chr3_+_142266113 | 0.73 |
ENSMUST00000106221.8
|
Gbp3
|
guanylate binding protein 3 |
| chr19_+_45549009 | 0.72 |
ENSMUST00000047057.9
|
Gm17018
|
predicted gene 17018 |
| chr10_-_94780695 | 0.71 |
ENSMUST00000099337.5
|
Plxnc1
|
plexin C1 |
| chr1_-_173318607 | 0.70 |
ENSMUST00000160565.4
|
Ifi206
|
interferon activated gene 206 |
| chr7_-_25488060 | 0.70 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr16_-_10603389 | 0.69 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chr16_-_35691914 | 0.69 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr8_+_62381115 | 0.69 |
ENSMUST00000154398.8
ENSMUST00000156980.8 ENSMUST00000093485.3 ENSMUST00000070631.15 |
Ddx60
|
DExD/H box helicase 60 |
| chr17_+_29879569 | 0.69 |
ENSMUST00000024816.13
ENSMUST00000235031.2 ENSMUST00000234911.2 |
Cmtr1
|
cap methyltransferase 1 |
| chrX_-_94488394 | 0.65 |
ENSMUST00000084535.6
|
Amer1
|
APC membrane recruitment 1 |
| chr3_-_94794256 | 0.64 |
ENSMUST00000005923.7
|
Psmb4
|
proteasome (prosome, macropain) subunit, beta type 4 |
| chr15_-_98626002 | 0.64 |
ENSMUST00000003445.8
|
Fkbp11
|
FK506 binding protein 11 |
| chr7_-_104114384 | 0.64 |
ENSMUST00000076922.6
|
Trim30a
|
tripartite motif-containing 30A |
| chr7_-_101667346 | 0.63 |
ENSMUST00000209844.2
ENSMUST00000211502.2 ENSMUST00000094134.5 |
Il18bp
|
interleukin 18 binding protein |
| chr7_-_126074222 | 0.63 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr1_-_52230062 | 0.62 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr15_-_98816012 | 0.62 |
ENSMUST00000023736.10
|
Lmbr1l
|
limb region 1 like |
| chr7_+_103893672 | 0.62 |
ENSMUST00000106848.8
|
Trim34a
|
tripartite motif-containing 34A |
| chr17_-_79190002 | 0.61 |
ENSMUST00000024884.5
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr14_-_78970160 | 0.61 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr9_-_78350486 | 0.61 |
ENSMUST00000070742.14
ENSMUST00000034898.14 |
Cgas
|
cyclic GMP-AMP synthase |
| chr11_+_58090382 | 0.61 |
ENSMUST00000035266.11
ENSMUST00000094169.11 ENSMUST00000168280.2 ENSMUST00000058704.9 |
Igtp
Irgm2
|
interferon gamma induced GTPase immunity-related GTPase family M member 2 |
| chr3_+_81839908 | 0.60 |
ENSMUST00000029649.3
|
Ctso
|
cathepsin O |
| chr8_+_72943455 | 0.60 |
ENSMUST00000072097.14
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr5_-_120915693 | 0.59 |
ENSMUST00000044833.9
|
Oas3
|
2'-5' oligoadenylate synthetase 3 |
| chr17_+_29879684 | 0.59 |
ENSMUST00000235014.2
ENSMUST00000130423.4 |
Cmtr1
|
cap methyltransferase 1 |
| chr14_+_55842002 | 0.58 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr1_+_153625243 | 0.58 |
ENSMUST00000182722.8
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr4_-_40239700 | 0.57 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr7_-_126074256 | 0.57 |
ENSMUST00000205440.2
ENSMUST00000032978.8 ENSMUST00000205340.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr15_-_77527470 | 0.56 |
ENSMUST00000181154.2
ENSMUST00000180949.8 ENSMUST00000166623.10 |
Apol11b
|
apolipoprotein L 11b |
| chr11_+_88890202 | 0.56 |
ENSMUST00000100627.9
ENSMUST00000107896.10 ENSMUST00000000284.7 |
Trim25
|
tripartite motif-containing 25 |
| chr1_+_52158721 | 0.55 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr1_+_153627692 | 0.55 |
ENSMUST00000183241.8
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr17_-_36501112 | 0.53 |
ENSMUST00000025312.13
ENSMUST00000102675.10 |
H2-T3
|
histocompatibility 2, T region locus 3 |
| chr2_-_51863203 | 0.53 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr2_-_25436884 | 0.52 |
ENSMUST00000114234.2
|
Traf2
|
TNF receptor-associated factor 2 |
| chr7_+_80510658 | 0.51 |
ENSMUST00000132163.8
ENSMUST00000205361.2 ENSMUST00000147125.2 |
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr7_-_126194097 | 0.51 |
ENSMUST00000058429.6
|
Il27
|
interleukin 27 |
| chr7_-_126073994 | 0.50 |
ENSMUST00000205733.2
ENSMUST00000205889.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr1_-_58544070 | 0.50 |
ENSMUST00000190695.2
|
Orc2
|
origin recognition complex, subunit 2 |
| chr3_-_104725853 | 0.49 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chr13_-_23894697 | 0.49 |
ENSMUST00000091707.13
ENSMUST00000006787.8 |
Hfe
|
homeostatic iron regulator |
| chr1_-_173859321 | 0.48 |
ENSMUST00000059226.7
|
Ifi205
|
interferon activated gene 205 |
| chr1_+_52158599 | 0.48 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr8_+_79236051 | 0.48 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr1_+_153625161 | 0.48 |
ENSMUST00000086209.10
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr6_-_39095144 | 0.48 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr11_+_72192455 | 0.47 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chr19_+_56385531 | 0.47 |
ENSMUST00000026062.10
|
Casp7
|
caspase 7 |
| chr5_+_107645626 | 0.47 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr11_-_121119864 | 0.47 |
ENSMUST00000137299.8
|
Cybc1
|
cytochrome b 245 chaperone 1 |
| chr13_+_120761861 | 0.46 |
ENSMUST00000225029.2
|
BC147527
|
cDNA sequence BC147527 |
| chr5_+_34494272 | 0.45 |
ENSMUST00000182047.2
|
Rnf4
|
ring finger protein 4 |
| chr3_-_104725581 | 0.45 |
ENSMUST00000168015.8
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr18_-_39622829 | 0.45 |
ENSMUST00000025300.13
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr13_-_14237958 | 0.44 |
ENSMUST00000223174.2
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr1_+_52158693 | 0.44 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr2_-_25436952 | 0.44 |
ENSMUST00000028311.13
|
Traf2
|
TNF receptor-associated factor 2 |
| chr3_+_89638044 | 0.43 |
ENSMUST00000029563.14
ENSMUST00000121094.8 ENSMUST00000118341.6 ENSMUST00000107405.6 |
Adar
|
adenosine deaminase, RNA-specific |
| chr13_-_23894828 | 0.43 |
ENSMUST00000091706.14
|
Hfe
|
homeostatic iron regulator |
| chr11_-_82882613 | 0.42 |
ENSMUST00000092840.11
ENSMUST00000038211.13 |
Slfn9
|
schlafen 9 |
| chr5_+_117495337 | 0.42 |
ENSMUST00000031309.16
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr2_-_65955338 | 0.42 |
ENSMUST00000028378.4
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr8_+_107877252 | 0.42 |
ENSMUST00000034400.5
|
Cyb5b
|
cytochrome b5 type B |
| chr11_-_100595019 | 0.40 |
ENSMUST00000017974.13
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr12_-_108859123 | 0.40 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr12_+_71021395 | 0.39 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr4_-_123466853 | 0.39 |
ENSMUST00000238555.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr11_+_29080733 | 0.39 |
ENSMUST00000020756.9
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr11_-_121120052 | 0.39 |
ENSMUST00000169393.8
ENSMUST00000106115.8 ENSMUST00000038709.14 ENSMUST00000147490.6 |
Cybc1
|
cytochrome b 245 chaperone 1 |
| chr4_-_156285247 | 0.39 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr6_+_90527762 | 0.38 |
ENSMUST00000130418.8
ENSMUST00000032175.11 ENSMUST00000203111.2 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr3_-_107667499 | 0.38 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr17_-_35081129 | 0.38 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr17_+_19724994 | 0.37 |
ENSMUST00000166081.3
ENSMUST00000231465.2 |
Vmn2r100
|
vomeronasal 2, receptor 100 |
| chr5_-_120950570 | 0.37 |
ENSMUST00000117193.8
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr4_-_155012534 | 0.37 |
ENSMUST00000219534.3
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr2_-_180883813 | 0.36 |
ENSMUST00000094203.11
ENSMUST00000108831.8 |
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr12_+_26519203 | 0.36 |
ENSMUST00000020969.5
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr3_-_104725535 | 0.36 |
ENSMUST00000002297.12
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr14_+_48683581 | 0.36 |
ENSMUST00000227440.2
ENSMUST00000124720.8 ENSMUST00000226422.2 ENSMUST00000226400.2 |
Tmem260
|
transmembrane protein 260 |
| chr3_+_127584251 | 0.35 |
ENSMUST00000164447.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr6_-_112673565 | 0.34 |
ENSMUST00000113182.8
ENSMUST00000113180.8 ENSMUST00000068487.12 ENSMUST00000077088.11 |
Rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr7_-_46445085 | 0.34 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr18_-_39623698 | 0.34 |
ENSMUST00000115567.8
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr9_+_96141317 | 0.33 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr1_+_85177316 | 0.33 |
ENSMUST00000161424.5
ENSMUST00000113402.4 |
Gm7609
|
predicted pseudogene 7609 |
| chr15_-_66684442 | 0.33 |
ENSMUST00000100572.10
|
Sla
|
src-like adaptor |
| chr19_+_36387123 | 0.33 |
ENSMUST00000225411.2
ENSMUST00000062389.6 |
Pcgf5
|
polycomb group ring finger 5 |
| chr4_+_138700195 | 0.33 |
ENSMUST00000123636.8
ENSMUST00000043042.10 ENSMUST00000050949.9 |
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr16_-_16176390 | 0.32 |
ENSMUST00000115749.3
ENSMUST00000230022.2 |
Dnm1l
|
dynamin 1-like |
| chr11_-_82911615 | 0.32 |
ENSMUST00000038141.15
ENSMUST00000092838.11 |
Slfn8
|
schlafen 8 |
| chr10_+_128106414 | 0.31 |
ENSMUST00000085708.3
ENSMUST00000105238.10 |
Stat2
|
signal transducer and activator of transcription 2 |
| chr9_+_108368032 | 0.31 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr3_+_20011201 | 0.31 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr1_+_127701901 | 0.31 |
ENSMUST00000112570.2
ENSMUST00000027587.15 |
Ccnt2
|
cyclin T2 |
| chr18_+_60345152 | 0.30 |
ENSMUST00000031549.6
|
Gm4951
|
predicted gene 4951 |
| chr1_-_58544105 | 0.29 |
ENSMUST00000191206.2
ENSMUST00000027198.12 |
Orc2
|
origin recognition complex, subunit 2 |
| chr2_-_132095146 | 0.29 |
ENSMUST00000028817.7
|
Pcna
|
proliferating cell nuclear antigen |
| chr2_-_120370333 | 0.29 |
ENSMUST00000171215.8
|
Zfp106
|
zinc finger protein 106 |
| chr2_-_180883746 | 0.29 |
ENSMUST00000121484.2
|
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr9_+_96141299 | 0.29 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr1_-_37535202 | 0.28 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr12_-_79054050 | 0.28 |
ENSMUST00000056660.13
ENSMUST00000174721.8 |
Tmem229b
|
transmembrane protein 229B |
| chr8_-_79547707 | 0.27 |
ENSMUST00000130325.8
ENSMUST00000051867.7 |
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr11_+_69737491 | 0.27 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr13_-_14237980 | 0.27 |
ENSMUST00000222687.2
ENSMUST00000221338.2 ENSMUST00000221713.2 ENSMUST00000170957.3 |
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr7_-_102214636 | 0.27 |
ENSMUST00000106913.3
ENSMUST00000033264.12 |
Trim21
|
tripartite motif-containing 21 |
| chr11_-_48817986 | 0.27 |
ENSMUST00000094476.9
|
Gm12185
|
predicted gene 12185 |
| chrX_+_7656251 | 0.26 |
ENSMUST00000140540.2
|
Gripap1
|
GRIP1 associated protein 1 |
| chr8_+_10056654 | 0.25 |
ENSMUST00000033892.9
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr5_-_90487583 | 0.25 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr3_-_14843512 | 0.25 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr1_+_175459735 | 0.25 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr17_-_35081456 | 0.24 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr15_+_81629258 | 0.24 |
ENSMUST00000109554.3
ENSMUST00000230946.2 |
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr15_-_103123711 | 0.24 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr10_-_98962187 | 0.24 |
ENSMUST00000060761.7
|
Phxr2
|
per-hexamer repeat gene 2 |
| chr7_-_99770653 | 0.24 |
ENSMUST00000208670.2
ENSMUST00000032969.14 |
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr19_+_30007910 | 0.23 |
ENSMUST00000025739.14
|
Uhrf2
|
ubiquitin-like, containing PHD and RING finger domains 2 |
| chr6_+_145067457 | 0.23 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr3_+_94861386 | 0.23 |
ENSMUST00000107260.9
ENSMUST00000142311.8 ENSMUST00000137088.8 ENSMUST00000152869.8 ENSMUST00000107254.8 ENSMUST00000107253.8 |
Rfx5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr2_-_51824656 | 0.22 |
ENSMUST00000165313.2
|
Rbm43
|
RNA binding motif protein 43 |
| chr2_-_51862941 | 0.22 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr13_-_30170031 | 0.22 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr11_+_72580823 | 0.21 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr18_+_69726055 | 0.21 |
ENSMUST00000114978.9
|
Tcf4
|
transcription factor 4 |
| chr7_+_58307930 | 0.21 |
ENSMUST00000168747.3
|
Atp10a
|
ATPase, class V, type 10A |
| chr13_-_103901010 | 0.21 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr2_-_91480096 | 0.21 |
ENSMUST00000099714.10
ENSMUST00000111333.2 |
Zfp408
|
zinc finger protein 408 |
| chr12_-_79027531 | 0.21 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr14_+_55815879 | 0.21 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr7_+_100742070 | 0.20 |
ENSMUST00000208439.2
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr3_+_55023594 | 0.20 |
ENSMUST00000146109.2
|
Spg20
|
spastic paraplegia 20, spartin (Troyer syndrome) homolog (human) |
| chr8_+_47192911 | 0.20 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr18_+_69726654 | 0.20 |
ENSMUST00000200921.4
|
Tcf4
|
transcription factor 4 |
| chr14_+_55815999 | 0.20 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr2_+_31462659 | 0.19 |
ENSMUST00000113482.8
|
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr11_+_4587733 | 0.19 |
ENSMUST00000070257.14
ENSMUST00000109930.3 |
Ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr3_+_145281941 | 0.19 |
ENSMUST00000199033.5
ENSMUST00000098534.9 ENSMUST00000200574.5 ENSMUST00000196413.5 ENSMUST00000197604.3 |
Znhit6
|
zinc finger, HIT type 6 |
| chr19_-_43741363 | 0.19 |
ENSMUST00000045562.6
|
Cox15
|
cytochrome c oxidase assembly protein 15 |
| chr7_+_130179063 | 0.18 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_-_58911902 | 0.18 |
ENSMUST00000134848.2
ENSMUST00000107557.9 ENSMUST00000149301.8 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr1_-_9818601 | 0.18 |
ENSMUST00000057438.7
|
Vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr2_-_36026614 | 0.18 |
ENSMUST00000122456.8
|
Rbm18
|
RNA binding motif protein 18 |
| chr2_+_180231038 | 0.18 |
ENSMUST00000029087.4
|
Ogfr
|
opioid growth factor receptor |
| chr4_-_42756489 | 0.18 |
ENSMUST00000140546.3
ENSMUST00000102957.6 |
Ccl19
|
chemokine (C-C motif) ligand 19 |
| chr2_+_31462780 | 0.17 |
ENSMUST00000137889.7
ENSMUST00000194386.6 ENSMUST00000055244.13 |
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr16_-_35759461 | 0.17 |
ENSMUST00000081933.14
ENSMUST00000114885.3 |
Dtx3l
|
deltex 3-like, E3 ubiquitin ligase |
| chr6_+_34840057 | 0.16 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr19_+_12438125 | 0.16 |
ENSMUST00000081035.9
|
Mpeg1
|
macrophage expressed gene 1 |
| chr2_-_131953359 | 0.16 |
ENSMUST00000128899.2
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr11_-_82911548 | 0.16 |
ENSMUST00000108152.9
|
Slfn8
|
schlafen 8 |
| chr15_+_74593041 | 0.15 |
ENSMUST00000070923.3
|
Them6
|
thioesterase superfamily member 6 |
| chr6_-_41752111 | 0.15 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr8_+_84703495 | 0.15 |
ENSMUST00000211558.2
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr1_-_75166994 | 0.15 |
ENSMUST00000189820.7
|
Atg9a
|
autophagy related 9A |
| chr9_+_44394080 | 0.14 |
ENSMUST00000220303.2
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.5 | 1.5 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.4 | 1.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 | 1.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 1.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 0.9 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.3 | 0.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 1.3 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 0.7 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 0.9 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.2 | 3.9 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.2 | 1.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.8 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 1.6 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.2 | 1.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.4 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.2 | 1.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.7 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) neutrophil clearance(GO:0097350) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.1 | 1.0 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.4 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.4 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.4 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.1 | 0.3 | GO:1900063 | mitochondrial membrane fission(GO:0090149) regulation of peroxisome organization(GO:1900063) |
| 0.1 | 5.1 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.6 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.5 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.6 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.1 | GO:0035697 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) |
| 0.1 | 0.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 3.3 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.1 | 0.9 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.6 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.5 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.3 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.4 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.3 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 1.6 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 0.6 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 3.4 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 0.6 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.2 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 1.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.9 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 | 0.3 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.7 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.3 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.7 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 1.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.2 | GO:0090308 | maintenance of DNA methylation(GO:0010216) regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 1.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.0 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 3.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.2 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 0.8 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.2 | 1.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.2 | 1.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 3.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.7 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.1 | 1.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 2.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.5 | 5.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 1.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.3 | 0.8 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.2 | 0.9 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 1.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 1.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 0.7 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.4 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.4 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.1 | 3.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 3.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.6 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0032139 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.5 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 2.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.1 | 0.6 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.2 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 3.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.6 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.5 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 4.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.0 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.7 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 3.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 1.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 3.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |