Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Stat4_Stat3_Stat5b
Z-value: 0.10
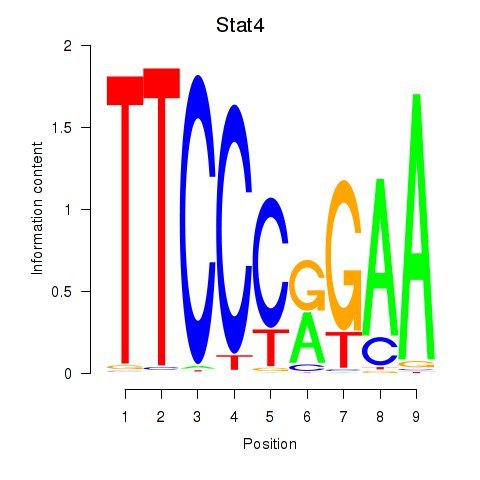
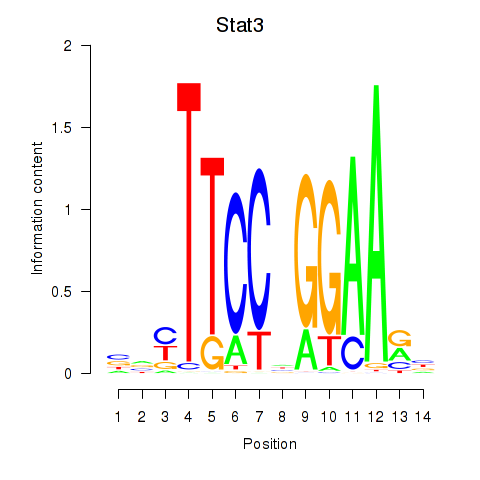
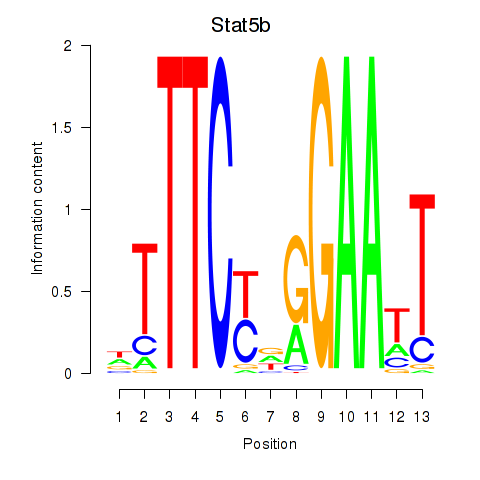
Transcription factors associated with Stat4_Stat3_Stat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat4
|
ENSMUSG00000062939.12 | signal transducer and activator of transcription 4 |
|
Stat3
|
ENSMUSG00000004040.17 | signal transducer and activator of transcription 3 |
|
Stat5b
|
ENSMUSG00000020919.12 | signal transducer and activator of transcription 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat3 | mm39_v1_chr11_-_100830366_100830388 | 0.65 | 2.4e-01 | Click! |
| Stat5b | mm39_v1_chr11_-_100713348_100713405 | 0.40 | 5.1e-01 | Click! |
| Stat4 | mm39_v1_chr1_+_52047368_52047413 | 0.20 | 7.5e-01 | Click! |
Activity profile of Stat4_Stat3_Stat5b motif
Sorted Z-values of Stat4_Stat3_Stat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_48977852 | 0.07 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr7_+_43086432 | 0.07 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr17_-_56428968 | 0.07 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr17_-_34406193 | 0.06 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr3_+_142202642 | 0.06 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr7_-_44785603 | 0.05 |
ENSMUST00000209467.2
|
Gm45713
|
predicted gene 45713 |
| chr1_+_34044940 | 0.05 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr1_+_134110142 | 0.05 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr7_+_43086554 | 0.05 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr9_+_20927271 | 0.04 |
ENSMUST00000086399.6
|
Icam1
|
intercellular adhesion molecule 1 |
| chr16_-_35691914 | 0.04 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr17_+_29309942 | 0.04 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr4_-_129534403 | 0.04 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr11_+_48977888 | 0.04 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr9_+_107173907 | 0.04 |
ENSMUST00000168260.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr11_-_53321242 | 0.04 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr8_-_112064783 | 0.04 |
ENSMUST00000056157.14
ENSMUST00000120432.3 |
Mlkl
|
mixed lineage kinase domain-like |
| chr13_-_42000958 | 0.03 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr11_-_117859997 | 0.03 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3 |
| chr12_-_110662256 | 0.03 |
ENSMUST00000149189.2
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr15_-_90563510 | 0.03 |
ENSMUST00000014777.9
ENSMUST00000064391.12 |
Cpne8
|
copine VIII |
| chr7_+_45204350 | 0.03 |
ENSMUST00000210300.2
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr6_-_136804728 | 0.03 |
ENSMUST00000146348.4
|
Wbp11
|
WW domain binding protein 11 |
| chr2_-_163592127 | 0.03 |
ENSMUST00000017841.4
|
Ada
|
adenosine deaminase |
| chr6_-_124888643 | 0.03 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr4_-_129534752 | 0.03 |
ENSMUST00000132217.8
ENSMUST00000130017.2 ENSMUST00000154105.8 |
Txlna
|
taxilin alpha |
| chr6_+_29468067 | 0.03 |
ENSMUST00000143101.4
ENSMUST00000149646.3 |
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr2_+_3425159 | 0.03 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr7_-_100505486 | 0.03 |
ENSMUST00000139604.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr7_-_101667346 | 0.03 |
ENSMUST00000209844.2
ENSMUST00000211502.2 ENSMUST00000094134.5 |
Il18bp
|
interleukin 18 binding protein |
| chr19_-_46950355 | 0.02 |
ENSMUST00000236501.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr17_+_34406523 | 0.02 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr6_+_34575435 | 0.02 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr16_+_17327076 | 0.02 |
ENSMUST00000232242.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr17_+_34406762 | 0.02 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr4_-_129534853 | 0.02 |
ENSMUST00000046425.16
ENSMUST00000133803.8 |
Txlna
|
taxilin alpha |
| chr4_+_11704438 | 0.02 |
ENSMUST00000108304.9
|
Gem
|
GTP binding protein (gene overexpressed in skeletal muscle) |
| chr6_-_129252396 | 0.02 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr10_-_17898938 | 0.02 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chr6_+_127423779 | 0.02 |
ENSMUST00000112191.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr8_-_112064393 | 0.02 |
ENSMUST00000145862.3
|
Mlkl
|
mixed lineage kinase domain-like |
| chr12_-_11315755 | 0.02 |
ENSMUST00000166117.4
ENSMUST00000219600.2 ENSMUST00000218487.2 |
Gen1
|
GEN1, Holliday junction 5' flap endonuclease |
| chr7_+_29607917 | 0.02 |
ENSMUST00000186475.2
|
Zfp383
|
zinc finger protein 383 |
| chr7_+_45204317 | 0.02 |
ENSMUST00000107752.12
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr14_-_99231754 | 0.02 |
ENSMUST00000081987.5
|
Rpl36a-ps1
|
ribosomal protein L36A, pseudogene 1 |
| chr1_+_134109888 | 0.02 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr13_-_67454476 | 0.02 |
ENSMUST00000049705.8
|
Zfp457
|
zinc finger protein 457 |
| chr9_+_119943916 | 0.02 |
ENSMUST00000135514.2
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr14_+_20344765 | 0.02 |
ENSMUST00000223663.2
ENSMUST00000022343.6 ENSMUST00000224066.2 ENSMUST00000223941.2 ENSMUST00000224311.2 |
Nudt13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr9_-_57460004 | 0.02 |
ENSMUST00000034856.15
|
Mpi
|
mannose phosphate isomerase |
| chr1_-_130656985 | 0.02 |
ENSMUST00000189534.7
|
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr12_-_112637998 | 0.02 |
ENSMUST00000128300.9
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr4_-_136613498 | 0.02 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr7_-_128063037 | 0.02 |
ENSMUST00000165023.8
|
Tial1
|
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr17_+_29898221 | 0.02 |
ENSMUST00000129864.2
|
Cmtr1
|
cap methyltransferase 1 |
| chr7_+_141056305 | 0.02 |
ENSMUST00000117634.2
|
Tspan4
|
tetraspanin 4 |
| chr18_-_42395207 | 0.02 |
ENSMUST00000097590.5
|
Lars
|
leucyl-tRNA synthetase |
| chr4_-_133746138 | 0.02 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr19_+_8870362 | 0.02 |
ENSMUST00000096249.7
|
Ints5
|
integrator complex subunit 5 |
| chr17_+_35598583 | 0.02 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr8_+_47192911 | 0.02 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr6_-_91450486 | 0.02 |
ENSMUST00000040835.9
|
Chchd4
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr16_-_90731394 | 0.02 |
ENSMUST00000142340.2
|
Cfap298
|
cilia and flagella associate protien 298 |
| chr15_+_101310283 | 0.02 |
ENSMUST00000068904.9
|
Krt7
|
keratin 7 |
| chr3_+_20011201 | 0.02 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr3_+_63203235 | 0.02 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr11_+_53661251 | 0.02 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr4_-_63965161 | 0.02 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr11_-_48762170 | 0.02 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr7_-_128062916 | 0.02 |
ENSMUST00000205835.2
|
Tial1
|
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr19_+_46611826 | 0.01 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr2_+_128942900 | 0.01 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr9_-_60594742 | 0.01 |
ENSMUST00000114032.8
ENSMUST00000166168.8 ENSMUST00000132366.2 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr1_-_85888729 | 0.01 |
ENSMUST00000086975.7
|
Gpr55
|
G protein-coupled receptor 55 |
| chr11_+_70861007 | 0.01 |
ENSMUST00000018593.10
|
Rpain
|
RPA interacting protein |
| chr17_-_13179187 | 0.01 |
ENSMUST00000159697.2
|
Acat2
|
acetyl-Coenzyme A acetyltransferase 2 |
| chr2_-_127324419 | 0.01 |
ENSMUST00000088538.6
|
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr8_+_39472981 | 0.01 |
ENSMUST00000239508.1
ENSMUST00000239509.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr17_-_52133594 | 0.01 |
ENSMUST00000129667.8
ENSMUST00000169480.8 ENSMUST00000148559.2 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr3_-_37180093 | 0.01 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr4_+_99812912 | 0.01 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr4_+_49521176 | 0.01 |
ENSMUST00000042964.13
ENSMUST00000107696.2 |
Zfp189
|
zinc finger protein 189 |
| chr5_+_134212836 | 0.01 |
ENSMUST00000016086.10
|
Gtf2ird2
|
GTF2I repeat domain containing 2 |
| chr5_-_34671236 | 0.01 |
ENSMUST00000114359.2
ENSMUST00000030991.14 ENSMUST00000087737.10 |
Tnip2
|
TNFAIP3 interacting protein 2 |
| chr17_+_26471889 | 0.01 |
ENSMUST00000114976.9
ENSMUST00000140427.8 ENSMUST00000119928.8 |
Luc7l
|
Luc7-like |
| chr11_-_78427061 | 0.01 |
ENSMUST00000017759.9
ENSMUST00000108277.3 |
Tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr2_-_84605764 | 0.01 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chrX_+_109857866 | 0.01 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr10_-_79911245 | 0.01 |
ENSMUST00000217972.2
|
Sbno2
|
strawberry notch 2 |
| chr11_+_48977495 | 0.01 |
ENSMUST00000152914.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr2_-_152256947 | 0.01 |
ENSMUST00000099207.5
|
Zcchc3
|
zinc finger, CCHC domain containing 3 |
| chr2_-_84605732 | 0.01 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr16_-_20245138 | 0.01 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr19_-_46033353 | 0.01 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr4_+_148125630 | 0.01 |
ENSMUST00000069604.15
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr10_-_39862622 | 0.01 |
ENSMUST00000170579.9
ENSMUST00000045524.7 ENSMUST00000073618.13 ENSMUST00000164566.8 |
Mfsd4b5
|
major facilitator superfamily domain containing 4B5 |
| chr15_-_55411560 | 0.01 |
ENSMUST00000165356.2
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr10_-_79936987 | 0.01 |
ENSMUST00000218630.2
|
Sbno2
|
strawberry notch 2 |
| chr2_+_128942919 | 0.01 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr16_+_21613068 | 0.01 |
ENSMUST00000211443.2
ENSMUST00000231300.2 ENSMUST00000209449.2 ENSMUST00000181780.9 ENSMUST00000209728.2 ENSMUST00000181960.3 ENSMUST00000209429.2 ENSMUST00000180830.3 ENSMUST00000231988.2 |
1300002E11Rik
Map3k13
|
RIKEN cDNA 1300002E11 gene mitogen-activated protein kinase kinase kinase 13 |
| chr6_-_129252323 | 0.01 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr15_-_77811935 | 0.01 |
ENSMUST00000174529.2
ENSMUST00000173631.8 |
Txn2
|
thioredoxin 2 |
| chr15_+_10224052 | 0.01 |
ENSMUST00000128450.8
ENSMUST00000148257.8 ENSMUST00000128921.8 |
Prlr
|
prolactin receptor |
| chr15_-_79430742 | 0.01 |
ENSMUST00000231053.2
ENSMUST00000229431.2 |
Ddx17
|
DEAD box helicase 17 |
| chr12_+_3476857 | 0.01 |
ENSMUST00000111215.10
ENSMUST00000092003.12 ENSMUST00000144247.9 ENSMUST00000153102.9 |
Asxl2
|
ASXL transcriptional regulator 2 |
| chr15_+_88960327 | 0.01 |
ENSMUST00000165690.2
|
Trabd
|
TraB domain containing |
| chr17_+_26471870 | 0.01 |
ENSMUST00000025023.15
|
Luc7l
|
Luc7-like |
| chrX_-_55643429 | 0.01 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr19_-_29025233 | 0.01 |
ENSMUST00000025696.5
|
Ak3
|
adenylate kinase 3 |
| chr14_-_70864448 | 0.01 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr2_-_65955338 | 0.01 |
ENSMUST00000028378.4
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr1_-_175520185 | 0.01 |
ENSMUST00000104984.4
ENSMUST00000209720.2 ENSMUST00000211489.2 ENSMUST00000210367.2 ENSMUST00000027809.8 |
Chml
Opn3
|
choroideremia-like opsin 3 |
| chr3_+_99048379 | 0.01 |
ENSMUST00000004343.7
|
Wars2
|
tryptophanyl tRNA synthetase 2 (mitochondrial) |
| chr7_-_128063354 | 0.01 |
ENSMUST00000106226.9
ENSMUST00000106228.8 |
Tial1
|
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr10_+_75009797 | 0.01 |
ENSMUST00000218465.2
|
Bcr
|
BCR activator of RhoGEF and GTPase |
| chr1_-_155688635 | 0.01 |
ENSMUST00000035325.15
|
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr11_+_53660834 | 0.01 |
ENSMUST00000108920.10
ENSMUST00000140866.9 ENSMUST00000108922.9 |
Irf1
|
interferon regulatory factor 1 |
| chrX_+_168468186 | 0.01 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr10_-_127147609 | 0.01 |
ENSMUST00000037290.12
ENSMUST00000171564.8 |
Mars1
|
methionine-tRNA synthetase 1 |
| chr19_-_10502468 | 0.01 |
ENSMUST00000025570.8
ENSMUST00000236455.2 |
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr7_+_29683373 | 0.01 |
ENSMUST00000148442.8
|
Zfp568
|
zinc finger protein 568 |
| chr15_+_102426742 | 0.01 |
ENSMUST00000100168.10
|
Tarbp2
|
TARBP2, RISC loading complex RNA binding subunit |
| chr13_-_33035150 | 0.01 |
ENSMUST00000091668.13
ENSMUST00000076352.8 |
Serpinb1a
|
serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| chr6_+_125108829 | 0.01 |
ENSMUST00000044200.11
ENSMUST00000204185.2 |
Nop2
|
NOP2 nucleolar protein |
| chr16_+_62674661 | 0.01 |
ENSMUST00000023629.9
|
Pros1
|
protein S (alpha) |
| chr17_+_28910302 | 0.01 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr7_-_99770653 | 0.01 |
ENSMUST00000208670.2
ENSMUST00000032969.14 |
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr18_-_34757653 | 0.01 |
ENSMUST00000003876.10
ENSMUST00000115766.8 ENSMUST00000097626.10 ENSMUST00000115765.2 |
Brd8
|
bromodomain containing 8 |
| chr19_-_47039261 | 0.01 |
ENSMUST00000026032.7
|
Pcgf6
|
polycomb group ring finger 6 |
| chr5_-_114518566 | 0.01 |
ENSMUST00000102581.11
|
Kctd10
|
potassium channel tetramerisation domain containing 10 |
| chr3_+_33853941 | 0.01 |
ENSMUST00000099153.10
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr14_+_55252911 | 0.01 |
ENSMUST00000022815.10
|
Ngdn
|
neuroguidin, EIF4E binding protein |
| chr9_+_45230370 | 0.01 |
ENSMUST00000034597.8
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr9_+_21914513 | 0.01 |
ENSMUST00000215795.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr3_-_127574167 | 0.01 |
ENSMUST00000029662.12
|
Alpk1
|
alpha-kinase 1 |
| chr9_+_48966868 | 0.01 |
ENSMUST00000034803.10
|
Zw10
|
zw10 kinetochore protein |
| chr15_+_100251030 | 0.01 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr9_-_21150100 | 0.01 |
ENSMUST00000164812.8
|
Keap1
|
kelch-like ECH-associated protein 1 |
| chr15_+_102427149 | 0.01 |
ENSMUST00000146756.8
ENSMUST00000142194.3 |
Tarbp2
|
TARBP2, RISC loading complex RNA binding subunit |
| chr5_+_52991351 | 0.01 |
ENSMUST00000031072.14
|
Anapc4
|
anaphase promoting complex subunit 4 |
| chr10_+_128139227 | 0.01 |
ENSMUST00000218315.2
ENSMUST00000219721.2 |
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr8_-_125296435 | 0.01 |
ENSMUST00000238882.2
ENSMUST00000063278.7 |
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr17_+_35345292 | 0.01 |
ENSMUST00000061859.7
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr16_-_56984137 | 0.01 |
ENSMUST00000231733.2
|
Nit2
|
nitrilase family, member 2 |
| chr8_-_83500740 | 0.01 |
ENSMUST00000034147.4
|
Zfp330
|
zinc finger protein 330 |
| chr7_+_27290969 | 0.01 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr12_+_83997382 | 0.01 |
ENSMUST00000053744.9
|
Riox1
|
ribosomal oxygenase 1 |
| chr3_+_99161070 | 0.01 |
ENSMUST00000029462.10
|
Tbx15
|
T-box 15 |
| chr10_+_80691099 | 0.01 |
ENSMUST00000035597.10
|
Sppl2b
|
signal peptide peptidase like 2B |
| chr16_-_91525485 | 0.01 |
ENSMUST00000231499.2
ENSMUST00000141664.9 ENSMUST00000123751.2 ENSMUST00000122254.8 ENSMUST00000114023.3 |
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr4_+_155896946 | 0.01 |
ENSMUST00000030944.11
|
Ccnl2
|
cyclin L2 |
| chr5_-_137608886 | 0.01 |
ENSMUST00000142675.8
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr12_+_52144511 | 0.01 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr8_+_106052970 | 0.01 |
ENSMUST00000015000.12
ENSMUST00000098453.9 |
Tmem208
|
transmembrane protein 208 |
| chr18_-_60860594 | 0.01 |
ENSMUST00000235795.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr10_+_80765900 | 0.01 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr11_-_70860778 | 0.01 |
ENSMUST00000108530.2
ENSMUST00000035283.11 ENSMUST00000108531.8 |
Nup88
|
nucleoporin 88 |
| chr11_-_68864666 | 0.01 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr3_+_94600863 | 0.01 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr6_+_3498382 | 0.01 |
ENSMUST00000001412.17
ENSMUST00000170873.10 ENSMUST00000164052.5 |
Vps50
|
VPS50 EARP/GARPII complex subunit |
| chr4_-_49521036 | 0.01 |
ENSMUST00000057829.4
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
| chr15_+_34238174 | 0.01 |
ENSMUST00000022867.5
ENSMUST00000226627.2 |
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr15_+_64689543 | 0.01 |
ENSMUST00000180105.2
|
Gm21798
|
predicted gene, 21798 |
| chr6_+_40605758 | 0.01 |
ENSMUST00000202636.4
ENSMUST00000201148.4 ENSMUST00000071535.10 |
Mgam
|
maltase-glucoamylase |
| chr7_-_23998735 | 0.01 |
ENSMUST00000145131.8
|
Zfp61
|
zinc finger protein 61 |
| chr18_-_70663209 | 0.01 |
ENSMUST00000161542.8
ENSMUST00000159389.8 |
Poli
|
polymerase (DNA directed), iota |
| chrX_-_133442596 | 0.01 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr9_-_21150350 | 0.01 |
ENSMUST00000049567.10
ENSMUST00000193982.2 |
Keap1
|
kelch-like ECH-associated protein 1 |
| chrX_+_47712676 | 0.01 |
ENSMUST00000177710.2
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr15_-_79430942 | 0.01 |
ENSMUST00000054014.9
ENSMUST00000229877.2 |
Ddx17
|
DEAD box helicase 17 |
| chr9_+_7272514 | 0.01 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr3_+_89958940 | 0.01 |
ENSMUST00000159064.8
|
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr14_+_55842002 | 0.01 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr7_+_29515485 | 0.01 |
ENSMUST00000178162.3
ENSMUST00000032796.14 |
Zfp790
|
zinc finger protein 790 |
| chr12_-_110669076 | 0.01 |
ENSMUST00000155242.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr8_-_22656428 | 0.01 |
ENSMUST00000033865.16
ENSMUST00000110730.10 |
Nek3
|
NIMA (never in mitosis gene a)-related expressed kinase 3 |
| chr1_-_155688551 | 0.01 |
ENSMUST00000194632.2
ENSMUST00000111764.8 |
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr8_+_95328558 | 0.01 |
ENSMUST00000060389.10
ENSMUST00000212729.2 ENSMUST00000121101.2 |
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr2_+_101716577 | 0.01 |
ENSMUST00000028584.8
|
Commd9
|
COMM domain containing 9 |
| chr16_-_4340920 | 0.01 |
ENSMUST00000090500.10
ENSMUST00000023161.8 |
Srl
|
sarcalumenin |
| chr11_+_115802828 | 0.01 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr17_+_34816826 | 0.01 |
ENSMUST00000015596.10
ENSMUST00000174496.9 ENSMUST00000173992.8 |
Ager
|
advanced glycosylation end product-specific receptor |
| chr13_+_67321219 | 0.01 |
ENSMUST00000021997.8
|
Rsl1
|
regulator of sex limited protein 1 |
| chr5_-_143255713 | 0.01 |
ENSMUST00000161448.8
|
Zfp316
|
zinc finger protein 316 |
| chr5_-_77555881 | 0.01 |
ENSMUST00000163898.6
ENSMUST00000046746.10 |
Igfbp7
|
insulin-like growth factor binding protein 7 |
| chr6_+_125044876 | 0.01 |
ENSMUST00000088292.7
|
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr5_+_110478558 | 0.01 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr13_-_96678987 | 0.01 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chr2_+_29780122 | 0.01 |
ENSMUST00000113762.8
ENSMUST00000113765.8 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr18_-_70663382 | 0.01 |
ENSMUST00000043286.15
|
Poli
|
polymerase (DNA directed), iota |
| chrX_+_47712614 | 0.01 |
ENSMUST00000114936.8
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr11_-_69786324 | 0.01 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr4_+_120523758 | 0.01 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr4_+_148686985 | 0.01 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chrX_-_133501677 | 0.01 |
ENSMUST00000239113.2
|
Gla
|
galactosidase, alpha |
| chr17_+_28945057 | 0.01 |
ENSMUST00000233003.2
|
Gm4356
|
predicted gene 4356 |
| chr13_+_34186346 | 0.01 |
ENSMUST00000021844.15
|
Ripk1
|
receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| chr18_+_50164043 | 0.01 |
ENSMUST00000145726.2
ENSMUST00000128377.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr9_+_65253374 | 0.01 |
ENSMUST00000048184.4
ENSMUST00000214433.2 |
Pdcd7
|
programmed cell death 7 |
| chr16_-_87292592 | 0.01 |
ENSMUST00000176750.2
ENSMUST00000175977.8 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat4_Stat3_Stat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |