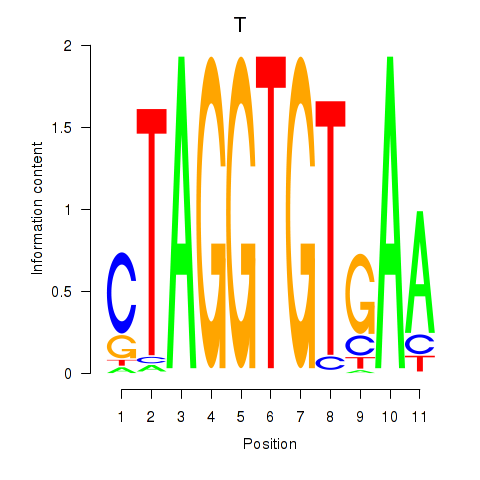
|
chr1_-_44258112
Show fit
|
0.43 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E
|
|
chr8_+_85449632
Show fit
|
0.35 |
ENSMUST00000098571.5
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene
|
|
chr9_-_43151179
Show fit
|
0.28 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog
|
|
chr14_+_50683002
Show fit
|
0.26 |
ENSMUST00000214792.2
|
Olfr740
|
olfactory receptor 740
|
|
chr5_-_142892457
Show fit
|
0.25 |
ENSMUST00000167721.8
ENSMUST00000163829.2
ENSMUST00000100497.11
|
Actb
|
actin, beta
|
|
chr11_-_69556904
Show fit
|
0.25 |
ENSMUST00000018918.12
|
Cd68
|
CD68 antigen
|
|
chr9_-_110818679
Show fit
|
0.24 |
ENSMUST00000084922.6
ENSMUST00000199891.2
|
Rtp3
|
receptor transporter protein 3
|
|
chr15_+_95688763
Show fit
|
0.23 |
ENSMUST00000227791.2
|
Ano6
|
anoctamin 6
|
|
chr7_+_5023552
Show fit
|
0.22 |
ENSMUST00000208728.2
ENSMUST00000085427.6
|
Ccdc106
Zfp865
|
coiled-coil domain containing 106
zinc finger protein 865
|
|
chr17_+_35286293
Show fit
|
0.21 |
ENSMUST00000173478.2
ENSMUST00000174876.2
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C
|
|
chr15_+_95688712
Show fit
|
0.21 |
ENSMUST00000071874.8
|
Ano6
|
anoctamin 6
|
|
chr19_-_40260060
Show fit
|
0.20 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin)
|
|
chr11_-_69556888
Show fit
|
0.19 |
ENSMUST00000108654.3
|
Cd68
|
CD68 antigen
|
|
chr16_+_27207722
Show fit
|
0.19 |
ENSMUST00000039443.14
ENSMUST00000096127.11
|
Ccdc50
|
coiled-coil domain containing 50
|
|
chr7_+_5023375
Show fit
|
0.18 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865
|
|
chr6_-_58195806
Show fit
|
0.18 |
ENSMUST00000228530.2
ENSMUST00000226666.2
|
Vmn1r27
|
vomeronasal 1 receptor 27
|
|
chr19_-_40260286
Show fit
|
0.18 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin)
|
|
chr12_-_111933328
Show fit
|
0.17 |
ENSMUST00000222874.2
ENSMUST00000223191.2
ENSMUST00000021719.7
|
Atp5mpl
|
ATP synthase membrane subunit 6.8PL
|
|
chr9_-_37525009
Show fit
|
0.17 |
ENSMUST00000002013.11
|
Spa17
|
sperm autoantigenic protein 17
|
|
chr9_-_57347366
Show fit
|
0.16 |
ENSMUST00000214144.2
ENSMUST00000085709.6
ENSMUST00000214624.2
ENSMUST00000215883.2
ENSMUST00000214339.2
ENSMUST00000215299.2
ENSMUST00000214166.2
ENSMUST00000214065.2
|
Ppcdc
|
phosphopantothenoylcysteine decarboxylase
|
|
chr2_+_164790139
Show fit
|
0.16 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9
|
|
chr15_+_59246134
Show fit
|
0.16 |
ENSMUST00000227173.2
ENSMUST00000079703.11
|
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase
|
|
chr3_-_103698391
Show fit
|
0.15 |
ENSMUST00000106845.9
ENSMUST00000029438.15
|
Hipk1
|
homeodomain interacting protein kinase 1
|
|
chr11_+_69804714
Show fit
|
0.14 |
ENSMUST00000072581.9
ENSMUST00000116358.8
|
Gps2
|
G protein pathway suppressor 2
|
|
chr11_-_94283792
Show fit
|
0.14 |
ENSMUST00000178136.8
ENSMUST00000021231.8
|
Abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3
|
|
chr14_+_80237691
Show fit
|
0.14 |
ENSMUST00000228749.2
ENSMUST00000088735.4
|
Olfm4
|
olfactomedin 4
|
|
chr11_-_100628979
Show fit
|
0.14 |
ENSMUST00000155500.2
ENSMUST00000107364.8
ENSMUST00000019317.12
|
Rab5c
|
RAB5C, member RAS oncogene family
|
|
chr8_+_121262528
Show fit
|
0.13 |
ENSMUST00000120493.8
|
Gse1
|
genetic suppressor element 1, coiled-coil protein
|
|
chr11_+_78427181
Show fit
|
0.13 |
ENSMUST00000128788.8
|
Ift20
|
intraflagellar transport 20
|
|
chrX_+_101048650
Show fit
|
0.13 |
ENSMUST00000144753.2
|
Nhsl2
|
NHS-like 2
|
|
chr15_+_79025523
Show fit
|
0.13 |
ENSMUST00000040077.8
|
Polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F
|
|
chr7_-_15361801
Show fit
|
0.13 |
ENSMUST00000086122.10
ENSMUST00000174443.8
|
Obox3
|
oocyte specific homeobox 3
|
|
chr11_-_54853729
Show fit
|
0.12 |
ENSMUST00000108885.8
ENSMUST00000102730.9
ENSMUST00000018482.13
ENSMUST00000108886.8
ENSMUST00000102731.8
|
Tnip1
|
TNFAIP3 interacting protein 1
|
|
chr8_+_70107131
Show fit
|
0.12 |
ENSMUST00000204285.3
|
Zfp964
|
zinc finger protein 964
|
|
chr19_+_23736205
Show fit
|
0.12 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1
|
|
chr18_-_36859732
Show fit
|
0.12 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen
|
|
chr9_+_50405817
Show fit
|
0.11 |
ENSMUST00000114474.8
ENSMUST00000188047.2
|
Plet1
|
placenta expressed transcript 1
|
|
chr17_+_84819260
Show fit
|
0.11 |
ENSMUST00000047206.7
|
Plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr7_+_24967094
Show fit
|
0.11 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor
|
|
chr10_+_130158737
Show fit
|
0.10 |
ENSMUST00000217702.2
ENSMUST00000042586.10
ENSMUST00000218605.2
|
Tespa1
|
thymocyte expressed, positive selection associated 1
|
|
chr5_+_105706713
Show fit
|
0.10 |
ENSMUST00000153754.2
|
Lrrc8c
|
leucine rich repeat containing 8 family, member C
|
|
chr10_+_23672842
Show fit
|
0.10 |
ENSMUST00000119597.8
ENSMUST00000179321.8
ENSMUST00000133289.2
|
Slc18b1
|
solute carrier family 18, subfamily B, member 1
|
|
chr6_-_53045546
Show fit
|
0.10 |
ENSMUST00000074541.6
|
Jazf1
|
JAZF zinc finger 1
|
|
chrX_-_142716200
Show fit
|
0.10 |
ENSMUST00000112851.8
ENSMUST00000112856.3
ENSMUST00000033642.10
|
Dcx
|
doublecortin
|
|
chr11_+_49094292
Show fit
|
0.10 |
ENSMUST00000150284.8
ENSMUST00000109197.8
ENSMUST00000151228.2
|
Zfp62
|
zinc finger protein 62
|
|
chr6_+_48624295
Show fit
|
0.09 |
ENSMUST00000078223.6
ENSMUST00000203509.2
|
Gimap8
|
GTPase, IMAP family member 8
|
|
chr1_-_156767196
Show fit
|
0.09 |
ENSMUST00000185198.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr13_-_38221045
Show fit
|
0.09 |
ENSMUST00000089840.5
|
Cage1
|
cancer antigen 1
|
|
chr1_+_88034556
Show fit
|
0.09 |
ENSMUST00000113137.2
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B
|
|
chr12_+_119356318
Show fit
|
0.08 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1
|
|
chr16_+_84631789
Show fit
|
0.08 |
ENSMUST00000114184.8
|
Gabpa
|
GA repeat binding protein, alpha
|
|
chr11_+_49094119
Show fit
|
0.08 |
ENSMUST00000109198.8
ENSMUST00000137061.9
|
Zfp62
|
zinc finger protein 62
|
|
chr18_-_12369351
Show fit
|
0.08 |
ENSMUST00000025279.6
|
Npc1
|
NPC intracellular cholesterol transporter 1
|
|
chr6_-_116084810
Show fit
|
0.08 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1
|
|
chr11_+_83328503
Show fit
|
0.08 |
ENSMUST00000037378.6
|
1700020L24Rik
|
RIKEN cDNA 1700020L24 gene
|
|
chrX_-_99669507
Show fit
|
0.08 |
ENSMUST00000059099.7
|
Pdzd11
|
PDZ domain containing 11
|
|
chr15_-_76004395
Show fit
|
0.08 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1
|
|
chr7_-_6733411
Show fit
|
0.07 |
ENSMUST00000239104.2
ENSMUST00000051209.11
|
Peg3
|
paternally expressed 3
|
|
chr9_-_40442669
Show fit
|
0.07 |
ENSMUST00000119373.9
|
Gramd1b
|
GRAM domain containing 1B
|
|
chr9_+_37524966
Show fit
|
0.07 |
ENSMUST00000215474.2
|
Siae
|
sialic acid acetylesterase
|
|
chr10_-_34294461
Show fit
|
0.07 |
ENSMUST00000213269.2
ENSMUST00000099973.4
ENSMUST00000105512.8
ENSMUST00000047885.14
|
Nt5dc1
|
5'-nucleotidase domain containing 1
|
|
chrX_-_161612373
Show fit
|
0.07 |
ENSMUST00000041370.11
ENSMUST00000112316.9
ENSMUST00000112315.2
|
Txlng
|
taxilin gamma
|
|
chr10_+_88295515
Show fit
|
0.07 |
ENSMUST00000125612.2
|
Sycp3
|
synaptonemal complex protein 3
|
|
chr14_+_6226418
Show fit
|
0.06 |
ENSMUST00000112625.9
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial
|
|
chr7_-_103832599
Show fit
|
0.06 |
ENSMUST00000216612.3
|
Olfr648
|
olfactory receptor 648
|
|
chr17_+_84013575
Show fit
|
0.06 |
ENSMUST00000112350.8
ENSMUST00000112349.9
ENSMUST00000112352.10
ENSMUST00000067826.15
|
Mta3
|
metastasis associated 3
|
|
chr6_-_116693849
Show fit
|
0.06 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72
|
|
chr4_+_155709275
Show fit
|
0.06 |
ENSMUST00000067081.10
ENSMUST00000105600.8
ENSMUST00000105598.2
|
Cdk11b
|
cyclin-dependent kinase 11B
|
|
chr12_-_87742525
Show fit
|
0.06 |
ENSMUST00000164517.3
|
Eif1ad19
|
eukaryotic translation initiation factor 1A domain containing 19
|
|
chr11_+_78427219
Show fit
|
0.06 |
ENSMUST00000050366.15
ENSMUST00000108275.2
|
Ift20
|
intraflagellar transport 20
|
|
chr10_-_12839995
Show fit
|
0.05 |
ENSMUST00000219727.2
ENSMUST00000163425.9
ENSMUST00000042861.7
ENSMUST00000218685.2
|
Stx11
|
syntaxin 11
|
|
chr11_-_86435579
Show fit
|
0.05 |
ENSMUST00000138810.3
ENSMUST00000058286.9
ENSMUST00000154617.8
|
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1
|
|
chr11_+_79551358
Show fit
|
0.05 |
ENSMUST00000155381.2
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II)
|
|
chr10_+_128158328
Show fit
|
0.05 |
ENSMUST00000219037.2
ENSMUST00000026446.4
|
Cnpy2
|
canopy FGF signaling regulator 2
|
|
chr1_+_88066086
Show fit
|
0.05 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A
|
|
chr16_+_84631956
Show fit
|
0.05 |
ENSMUST00000009120.8
|
Gabpa
|
GA repeat binding protein, alpha
|
|
chr8_+_124133832
Show fit
|
0.05 |
ENSMUST00000098324.4
|
Mc1r
|
melanocortin 1 receptor
|
|
chr19_+_28941292
Show fit
|
0.05 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6
|
|
chr6_-_94677118
Show fit
|
0.05 |
ENSMUST00000101126.3
ENSMUST00000032105.11
|
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr13_-_81781238
Show fit
|
0.05 |
ENSMUST00000126444.8
ENSMUST00000128585.9
ENSMUST00000146749.2
ENSMUST00000095585.11
|
Adgrv1
|
adhesion G protein-coupled receptor V1
|
|
chr4_+_146533480
Show fit
|
0.04 |
ENSMUST00000105733.3
|
Zfp992
|
zinc finger protein 992
|
|
chr7_+_110376859
Show fit
|
0.04 |
ENSMUST00000148292.2
|
Ampd3
|
adenosine monophosphate deaminase 3
|
|
chr9_-_107166543
Show fit
|
0.04 |
ENSMUST00000192054.2
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr6_-_24956296
Show fit
|
0.04 |
ENSMUST00000127247.4
|
Tmem229a
|
transmembrane protein 229A
|
|
chr6_+_72281587
Show fit
|
0.04 |
ENSMUST00000183018.8
ENSMUST00000182014.10
|
Sftpb
|
surfactant associated protein B
|
|
chr9_-_69668204
Show fit
|
0.04 |
ENSMUST00000071281.5
|
Foxb1
|
forkhead box B1
|
|
chr3_-_103698853
Show fit
|
0.04 |
ENSMUST00000118317.8
|
Hipk1
|
homeodomain interacting protein kinase 1
|
|
chr2_+_32340459
Show fit
|
0.04 |
ENSMUST00000048431.3
|
Naif1
|
nuclear apoptosis inducing factor 1
|
|
chr15_-_59245998
Show fit
|
0.04 |
ENSMUST00000022976.6
|
Washc5
|
WASH complex subunit 5
|
|
chr15_+_79776597
Show fit
|
0.03 |
ENSMUST00000175714.8
ENSMUST00000109620.10
ENSMUST00000165537.8
ENSMUST00000175752.8
ENSMUST00000176325.8
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3
|
|
chr8_-_34333573
Show fit
|
0.03 |
ENSMUST00000183062.2
|
Rbpms
|
RNA binding protein gene with multiple splicing
|
|
chr9_-_95727267
Show fit
|
0.03 |
ENSMUST00000093800.9
|
Pls1
|
plastin 1 (I-isoform)
|
|
chr9_-_20657643
Show fit
|
0.03 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2
|
|
chr9_-_15023396
Show fit
|
0.03 |
ENSMUST00000159985.2
|
Hephl1
|
hephaestin-like 1
|
|
chr2_-_180844582
Show fit
|
0.03 |
ENSMUST00000016511.6
|
Ptk6
|
PTK6 protein tyrosine kinase 6
|
|
chr6_-_122587005
Show fit
|
0.03 |
ENSMUST00000032211.5
|
Gdf3
|
growth differentiation factor 3
|
|
chr8_+_107662352
Show fit
|
0.03 |
ENSMUST00000212524.2
ENSMUST00000047425.5
|
Sntb2
|
syntrophin, basic 2
|
|
chr3_+_40905066
Show fit
|
0.03 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B
|
|
chr7_+_126028225
Show fit
|
0.03 |
ENSMUST00000106407.9
|
Rabep2
|
rabaptin, RAB GTPase binding effector protein 2
|
|
chr1_-_71454041
Show fit
|
0.03 |
ENSMUST00000087268.7
|
Abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr1_-_155908695
Show fit
|
0.03 |
ENSMUST00000136397.8
|
Tor1aip1
|
torsin A interacting protein 1
|
|
chr9_+_121548893
Show fit
|
0.03 |
ENSMUST00000182337.2
|
Nktr
|
natural killer tumor recognition sequence
|
|
chr8_-_83129832
Show fit
|
0.03 |
ENSMUST00000034148.7
|
Il15
|
interleukin 15
|
|
chr4_+_155648256
Show fit
|
0.03 |
ENSMUST00000143840.2
ENSMUST00000146080.8
|
Nadk
|
NAD kinase
|
|
chr3_+_40905216
Show fit
|
0.03 |
ENSMUST00000191872.6
ENSMUST00000200432.2
|
Larp1b
|
La ribonucleoprotein domain family, member 1B
|
|
chr7_+_26854507
Show fit
|
0.02 |
ENSMUST00000108385.3
|
Cyp2t4
|
cytochrome P450, family 2, subfamily t, polypeptide 4
|
|
chr14_-_119162736
Show fit
|
0.02 |
ENSMUST00000004055.10
|
Dzip1
|
DAZ interacting protein 1
|
|
chr2_-_125993887
Show fit
|
0.02 |
ENSMUST00000110448.3
ENSMUST00000110446.9
|
Fam227b
|
family with sequence similarity 227, member B
|
|
chr10_+_97528915
Show fit
|
0.02 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1
|
|
chr18_+_38429858
Show fit
|
0.02 |
ENSMUST00000171461.3
ENSMUST00000235811.2
|
Rnf14
|
ring finger protein 14
|
|
chr17_-_78991691
Show fit
|
0.02 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein
|
|
chr13_+_38220971
Show fit
|
0.02 |
ENSMUST00000021866.10
|
Riok1
|
RIO kinase 1
|
|
chr7_-_79492091
Show fit
|
0.02 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase
|
|
chr4_+_138559156
Show fit
|
0.02 |
ENSMUST00000077582.14
|
Pla2g2a
|
phospholipase A2, group IIA (platelets, synovial fluid)
|
|
chr1_+_181404124
Show fit
|
0.02 |
ENSMUST00000208001.2
|
Dnah14
|
dynein, axonemal, heavy chain 14
|
|
chr13_-_38220899
Show fit
|
0.02 |
ENSMUST00000110233.8
ENSMUST00000074969.11
ENSMUST00000131066.2
|
Cage1
|
cancer antigen 1
|
|
chr10_+_21854540
Show fit
|
0.02 |
ENSMUST00000142174.8
ENSMUST00000164659.8
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_26386609
Show fit
|
0.02 |
ENSMUST00000193603.6
|
Nlgn1
|
neuroligin 1
|
|
chr12_-_113589576
Show fit
|
0.02 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6
|
|
chr5_+_100054110
Show fit
|
0.02 |
ENSMUST00000198837.3
|
Vamp9
|
vesicle-associated membrane protein 9
|
|
chr2_+_74528071
Show fit
|
0.02 |
ENSMUST00000059272.10
|
Hoxd9
|
homeobox D9
|
|
chr8_-_73246435
Show fit
|
0.01 |
ENSMUST00000152080.8
|
Slc35e1
|
solute carrier family 35, member E1
|
|
chr3_-_107851021
Show fit
|
0.01 |
ENSMUST00000106684.8
ENSMUST00000106685.9
|
Gstm6
|
glutathione S-transferase, mu 6
|
|
chr12_-_113625906
Show fit
|
0.01 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9
|
|
chr14_+_13803477
Show fit
|
0.01 |
ENSMUST00000102996.4
|
Cfap20dc
|
CFAP20 domain containing
|
|
chr9_+_86349226
Show fit
|
0.01 |
ENSMUST00000188675.7
|
Dop1a
|
DOP1 leucine zipper like protein A
|
|
chr19_-_45619559
Show fit
|
0.01 |
ENSMUST00000160718.9
|
Fbxw4
|
F-box and WD-40 domain protein 4
|
|
chr2_+_181134977
Show fit
|
0.01 |
ENSMUST00000069649.9
|
Abhd16b
|
abhydrolase domain containing 16B
|
|
chr6_+_90310252
Show fit
|
0.01 |
ENSMUST00000046128.12
ENSMUST00000164761.6
|
Uroc1
|
urocanase domain containing 1
|
|
chr10_-_95678748
Show fit
|
0.01 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543
|
|
chr15_+_9335636
Show fit
|
0.01 |
ENSMUST00000072403.7
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2
|
|
chr8_+_61085853
Show fit
|
0.01 |
ENSMUST00000161421.2
|
Mfap3l
|
microfibrillar-associated protein 3-like
|
|
chr11_+_78068931
Show fit
|
0.01 |
ENSMUST00000147819.8
|
Tlcd1
|
TLC domain containing 1
|
|
chr10_-_95678786
Show fit
|
0.01 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543
|
|
chr7_-_30335277
Show fit
|
0.01 |
ENSMUST00000108147.3
|
Etv2
|
ets variant 2
|
|
chr7_-_89166781
Show fit
|
0.01 |
ENSMUST00000041761.7
|
Prss23
|
protease, serine 23
|
|
chr9_-_9239056
Show fit
|
0.01 |
ENSMUST00000093893.12
|
Arhgap42
|
Rho GTPase activating protein 42
|
|
chr1_-_86317066
Show fit
|
0.01 |
ENSMUST00000212058.2
|
Nmur1
|
neuromedin U receptor 1
|
|
chr14_-_40791151
Show fit
|
0.01 |
ENSMUST00000022316.6
|
Dydc2
|
DPY30 domain containing 2
|
|
chr7_-_79443536
Show fit
|
0.01 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1
|
|
chr7_-_119694400
Show fit
|
0.01 |
ENSMUST00000209154.3
ENSMUST00000046993.4
|
Dnah3
|
dynein, axonemal, heavy chain 3
|
|
chr16_-_44153498
Show fit
|
0.01 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1
|
|
chr4_+_52964547
Show fit
|
0.01 |
ENSMUST00000215010.2
ENSMUST00000215127.2
|
Olfr270
|
olfactory receptor 270
|
|
chr16_-_55755208
Show fit
|
0.01 |
ENSMUST00000121129.8
ENSMUST00000023270.14
|
Cep97
|
centrosomal protein 97
|
|
chr15_-_48655329
Show fit
|
0.01 |
ENSMUST00000160658.8
ENSMUST00000100670.10
ENSMUST00000162830.8
|
Csmd3
|
CUB and Sushi multiple domains 3
|
|
chr15_-_82291372
Show fit
|
0.01 |
ENSMUST00000230198.2
ENSMUST00000230248.2
ENSMUST00000072776.5
ENSMUST00000229911.2
|
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10
|
|
chrX_-_138683102
Show fit
|
0.01 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1
|
|
chr4_-_118693476
Show fit
|
0.01 |
ENSMUST00000214477.2
|
Olfr1333
|
olfactory receptor 1333
|
|
chr5_+_73563418
Show fit
|
0.01 |
ENSMUST00000031040.13
ENSMUST00000065543.8
|
Cwh43
|
cell wall biogenesis 43 C-terminal homolog
|
|
chr16_-_44566700
Show fit
|
0.01 |
ENSMUST00000023348.11
ENSMUST00000162512.8
|
Gtpbp8
|
GTP-binding protein 8 (putative)
|
|
chr8_+_114369838
Show fit
|
0.01 |
ENSMUST00000095173.3
ENSMUST00000034219.12
ENSMUST00000212269.2
|
Syce1l
|
synaptonemal complex central element protein 1 like
|
|
chr2_+_150942528
Show fit
|
0.01 |
ENSMUST00000178211.2
|
Gm14147
|
predicted gene 14147
|
|
chr15_-_98155173
Show fit
|
0.01 |
ENSMUST00000060855.7
|
H1f7
|
H1.7 linker histone
|
|
chr16_-_44153288
Show fit
|
0.01 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1
|
|
chr3_-_126918491
Show fit
|
0.01 |
ENSMUST00000238781.2
|
Ank2
|
ankyrin 2, brain
|
|
chr12_-_113542610
Show fit
|
0.01 |
ENSMUST00000195468.6
ENSMUST00000103442.3
|
Ighv5-2
|
immunoglobulin heavy variable 5-2
|
|
chr4_-_139974062
Show fit
|
0.01 |
ENSMUST00000039331.9
|
Igsf21
|
immunoglobulin superfamily, member 21
|
|
chr3_-_121076745
Show fit
|
0.01 |
ENSMUST00000135818.8
ENSMUST00000137234.2
|
Tlcd4
|
TLC domain containing 4
|
|
chr3_+_108987779
Show fit
|
0.01 |
ENSMUST00000029478.5
|
Slc25a54
|
solute carrier family 25, member 54
|
|
chr7_+_14393658
Show fit
|
0.01 |
ENSMUST00000069740.14
ENSMUST00000183424.2
|
Obox7
|
oocyte specific homeobox 7
|
|
chr15_-_60793115
Show fit
|
0.01 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein
|
|
chr4_-_129132963
Show fit
|
0.01 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080
|
|
chr16_-_32617783
Show fit
|
0.01 |
ENSMUST00000115116.8
ENSMUST00000041123.9
|
Muc20
|
mucin 20
|
|
chr4_-_41731142
Show fit
|
0.01 |
ENSMUST00000171251.8
|
Arid3c
|
AT rich interactive domain 3C (BRIGHT-like)
|
|
chr7_-_107446697
Show fit
|
0.01 |
ENSMUST00000220193.2
|
Olfr470
|
olfactory receptor 470
|
|
chr14_-_75829389
Show fit
|
0.01 |
ENSMUST00000165569.3
ENSMUST00000035243.5
|
Cby2
|
chibby family member 2
|
|
chr3_-_158267771
Show fit
|
0.01 |
ENSMUST00000199890.5
ENSMUST00000238317.3
ENSMUST00000200137.5
ENSMUST00000106044.6
|
Lrrc7
|
leucine rich repeat containing 7
|
|
chr8_-_70664901
Show fit
|
0.00 |
ENSMUST00000063788.8
ENSMUST00000110127.8
|
Slc25a42
|
solute carrier family 25, member 42
|
|
chr7_+_140284078
Show fit
|
0.00 |
ENSMUST00000080681.4
|
Olfr541
|
olfactory receptor 541
|
|
chr9_-_99592058
Show fit
|
0.00 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18
|
|
chr6_-_119444157
Show fit
|
0.00 |
ENSMUST00000118120.8
|
Wnt5b
|
wingless-type MMTV integration site family, member 5B
|
|
chr7_+_14393421
Show fit
|
0.00 |
ENSMUST00000183788.8
|
Obox7
|
oocyte specific homeobox 7
|
|
chr15_-_101759212
Show fit
|
0.00 |
ENSMUST00000023790.5
|
Krt1
|
keratin 1
|
|
chr9_-_111519382
Show fit
|
0.00 |
ENSMUST00000035083.8
|
Stac
|
src homology three (SH3) and cysteine rich domain
|
|
chr19_+_37686240
Show fit
|
0.00 |
ENSMUST00000025946.7
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1
|
|
chr17_+_25588241
Show fit
|
0.00 |
ENSMUST00000160377.8
ENSMUST00000160485.8
|
Tpsg1
|
tryptase gamma 1
|
|
chr9_-_78263025
Show fit
|
0.00 |
ENSMUST00000125479.8
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2)
|
|
chr6_-_139987135
Show fit
|
0.00 |
ENSMUST00000032356.13
|
Plcz1
|
phospholipase C, zeta 1
|
|
chr10_-_13053733
Show fit
|
0.00 |
ENSMUST00000019954.6
|
Zc2hc1b
|
zinc finger, C2HC-type containing 1B
|
|
chr10_+_127541039
Show fit
|
0.00 |
ENSMUST00000079590.7
|
Myo1a
|
myosin IA
|
|
chr7_-_130931235
Show fit
|
0.00 |
ENSMUST00000188899.2
|
Fam24b
|
family with sequence similarity 24 member B
|
|
chr7_+_15484056
Show fit
|
0.00 |
ENSMUST00000173455.8
|
Obox5
|
oocyte specific homeobox 5
|
|
chr5_-_31250817
Show fit
|
0.00 |
ENSMUST00000031037.14
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr15_-_101588714
Show fit
|
0.00 |
ENSMUST00000023786.7
|
Krt6b
|
keratin 6B
|
|
chrX_-_149440388
Show fit
|
0.00 |
ENSMUST00000151403.9
ENSMUST00000087253.11
ENSMUST00000112709.8
ENSMUST00000163969.8
ENSMUST00000087258.10
|
Tro
|
trophinin
|
|
chr1_-_13197387
Show fit
|
0.00 |
ENSMUST00000047577.7
|
Prdm14
|
PR domain containing 14
|
|
chr2_-_17395765
Show fit
|
0.00 |
ENSMUST00000177966.2
|
Nebl
|
nebulette
|
|
chr2_+_129965195
Show fit
|
0.00 |
ENSMUST00000028888.5
|
Tgm6
|
transglutaminase 6
|
|
chr14_-_42461286
Show fit
|
0.00 |
ENSMUST00000163102.2
|
Gm3633
|
predicted gene 3633
|
|
chr10_+_88295431
Show fit
|
0.00 |
ENSMUST00000020252.10
|
Sycp3
|
synaptonemal complex protein 3
|
|
chr15_-_101602734
Show fit
|
0.00 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A
|
|
chr9_-_112014837
Show fit
|
0.00 |
ENSMUST00000162065.8
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21
|
|
chr14_-_21790463
Show fit
|
0.00 |
ENSMUST00000120984.9
|
Dusp13
|
dual specificity phosphatase 13
|
|
chr2_-_27138347
Show fit
|
0.00 |
ENSMUST00000139312.8
|
Sardh
|
sarcosine dehydrogenase
|
|
chr9_-_99592116
Show fit
|
0.00 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18
|
|
chr14_-_24294933
Show fit
|
0.00 |
ENSMUST00000169880.3
|
Dlg5
|
discs large MAGUK scaffold protein 5
|
|
chrX_-_149440362
Show fit
|
0.00 |
ENSMUST00000148604.2
|
Tro
|
trophinin
|
|
chr8_+_46338557
Show fit
|
0.00 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3
|
|
chr8_+_3520841
Show fit
|
0.00 |
ENSMUST00000047265.13
|
Tex45
|
testis expressed 45
|
|
chr6_-_141954471
Show fit
|
0.00 |
ENSMUST00000111832.3
|
Gm6614
|
predicted gene 6614
|