Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Tbp
Z-value: 6.36
Transcription factors associated with Tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbp
|
ENSMUSG00000014767.18 | TATA box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbp | mm39_v1_chr17_+_15720150_15720222 | 0.55 | 3.3e-01 | Click! |
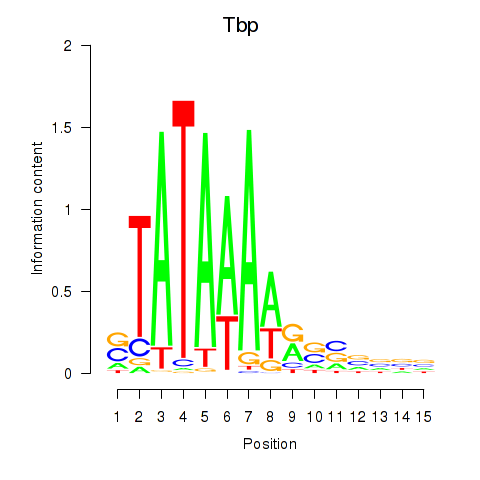
Activity profile of Tbp motif
Sorted Z-values of Tbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23765546 | 17.50 |
ENSMUST00000102968.3
|
H4c4
|
H4 clustered histone 4 |
| chr13_-_24118139 | 16.14 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr13_+_23936250 | 14.57 |
ENSMUST00000091703.3
|
H3c2
|
H3 clustered histone 2 |
| chr13_-_23882437 | 13.87 |
ENSMUST00000102967.3
|
H4c3
|
H4 clustered histone 3 |
| chr13_-_23806530 | 13.30 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr6_-_72935382 | 12.55 |
ENSMUST00000144337.2
|
Tmsb10
|
thymosin, beta 10 |
| chr13_+_21901791 | 12.35 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr3_+_96154001 | 12.27 |
ENSMUST00000176059.2
ENSMUST00000177796.2 |
H3c14
|
H3 clustered histone 14 |
| chr13_-_21934675 | 12.26 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr3_+_96175970 | 11.41 |
ENSMUST00000098843.3
|
H3c13
|
H3 clustered histone 13 |
| chr13_+_23728222 | 11.31 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr13_-_21967540 | 10.60 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11 |
| chr13_+_23947641 | 10.38 |
ENSMUST00000055770.4
|
H1f1
|
H1.1 linker histone, cluster member |
| chr13_+_23759930 | 10.19 |
ENSMUST00000105105.4
|
H3c4
|
H3 clustered histone 4 |
| chr13_+_22017906 | 10.15 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24 |
| chr13_+_21971631 | 10.11 |
ENSMUST00000110473.3
ENSMUST00000102982.2 |
H2bc22
|
H2B clustered histone 22 |
| chr14_-_51045182 | 9.71 |
ENSMUST00000227614.2
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr13_-_23946359 | 9.16 |
ENSMUST00000091701.3
|
H3c1
|
H3 clustered histone 1 |
| chr13_-_23929490 | 9.07 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr13_+_23715220 | 8.64 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr13_+_23868175 | 7.96 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr3_+_96152813 | 7.91 |
ENSMUST00000078756.7
ENSMUST00000090779.4 |
H2ac18
Gm20634
|
H2A clustered histone 18 predicted gene 20634 |
| chr13_+_21919225 | 7.90 |
ENSMUST00000087714.6
|
H4c11
|
H4 clustered histone 11 |
| chr13_+_21900554 | 6.85 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr17_+_36227869 | 6.15 |
ENSMUST00000151664.8
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr13_-_23867924 | 5.93 |
ENSMUST00000171127.4
|
H2ac6
|
H2A clustered histone 6 |
| chr6_-_72935468 | 5.88 |
ENSMUST00000114050.8
|
Tmsb10
|
thymosin, beta 10 |
| chr13_-_22017677 | 5.81 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr13_-_21971388 | 5.53 |
ENSMUST00000091751.3
|
H2ac22
|
H2A clustered histone 22 |
| chr3_-_20296337 | 5.50 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr7_-_15993110 | 5.23 |
ENSMUST00000168818.2
|
C5ar1
|
complement component 5a receptor 1 |
| chr2_+_129854256 | 3.94 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr6_-_72935171 | 3.64 |
ENSMUST00000114049.2
|
Tmsb10
|
thymosin, beta 10 |
| chr1_-_171023798 | 3.32 |
ENSMUST00000111332.2
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr8_+_73072877 | 3.08 |
ENSMUST00000067912.8
|
Klf2
|
Kruppel-like factor 2 (lung) |
| chr13_-_22225527 | 3.07 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9 |
| chr13_-_23755374 | 3.04 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr9_+_7558449 | 2.94 |
ENSMUST00000018765.4
|
Mmp8
|
matrix metallopeptidase 8 |
| chr17_-_34043320 | 2.81 |
ENSMUST00000173879.8
ENSMUST00000166693.3 ENSMUST00000173019.8 |
Rps28
|
ribosomal protein S28 |
| chr15_-_76116245 | 2.80 |
ENSMUST00000167754.8
|
Plec
|
plectin |
| chr15_+_3300249 | 2.74 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr10_-_128462616 | 2.53 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
| chr13_+_23758555 | 2.48 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr2_+_111144362 | 2.45 |
ENSMUST00000219291.2
|
Olfr1280
|
olfactory receptor 1280 |
| chr2_+_152578164 | 2.42 |
ENSMUST00000038368.9
ENSMUST00000109824.2 |
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chr11_-_4897991 | 2.38 |
ENSMUST00000093369.5
|
Nefh
|
neurofilament, heavy polypeptide |
| chr7_+_30252687 | 2.30 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr9_+_57818243 | 2.21 |
ENSMUST00000216925.2
ENSMUST00000163329.2 ENSMUST00000213654.2 ENSMUST00000217132.2 ENSMUST00000216841.2 ENSMUST00000214086.2 |
Ubl7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chr17_+_18269686 | 1.81 |
ENSMUST00000176802.3
|
Vmn2r124
|
vomeronasal 2, receptor 124 |
| chr18_-_78166595 | 1.78 |
ENSMUST00000091813.12
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr5_-_125467063 | 1.74 |
ENSMUST00000136312.2
|
Ubc
|
ubiquitin C |
| chr17_+_36227433 | 1.73 |
ENSMUST00000087211.9
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr15_-_102154874 | 1.66 |
ENSMUST00000063339.14
|
Rarg
|
retinoic acid receptor, gamma |
| chr1_-_89942299 | 1.65 |
ENSMUST00000086882.8
ENSMUST00000097656.10 |
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr11_-_120604551 | 1.56 |
ENSMUST00000106154.8
ENSMUST00000106155.4 ENSMUST00000055424.13 ENSMUST00000026137.8 |
Cenpx
|
centromere protein X |
| chr2_-_51039112 | 1.51 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr10_+_51367052 | 1.48 |
ENSMUST00000217705.2
ENSMUST00000078778.5 ENSMUST00000220182.2 ENSMUST00000220226.2 |
Lilrb4a
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4A |
| chrX_+_158623460 | 1.48 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr8_-_3798941 | 1.47 |
ENSMUST00000012847.3
|
Cd209a
|
CD209a antigen |
| chr8_-_3798922 | 1.47 |
ENSMUST00000208960.2
ENSMUST00000207979.2 |
Cd209a
|
CD209a antigen |
| chr4_-_123507494 | 1.45 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr16_-_55642491 | 1.42 |
ENSMUST00000114458.8
|
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr4_+_40269563 | 1.42 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr16_-_55659194 | 1.38 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr7_-_45109262 | 1.34 |
ENSMUST00000094434.13
|
Ftl1
|
ferritin light polypeptide 1 |
| chr2_+_172090412 | 1.32 |
ENSMUST00000038532.2
|
Mc3r
|
melanocortin 3 receptor |
| chr1_+_93918231 | 1.20 |
ENSMUST00000097632.5
|
Gal3st2c
|
galactose-3-O-sulfotransferase 2C |
| chr11_-_73215442 | 1.19 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr19_-_3464447 | 1.18 |
ENSMUST00000025842.8
ENSMUST00000237521.2 |
Gal
|
galanin and GMAP prepropeptide |
| chrX_+_102400061 | 1.17 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr5_-_125467240 | 1.15 |
ENSMUST00000156249.2
|
Ubc
|
ubiquitin C |
| chr2_+_90927053 | 1.13 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr2_-_166904625 | 1.10 |
ENSMUST00000128676.2
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr5_-_91550853 | 1.07 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr5_-_71253107 | 1.04 |
ENSMUST00000197284.5
|
Gabra2
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
| chr7_+_108209994 | 1.04 |
ENSMUST00000209296.3
|
Olfr506
|
olfactory receptor 506 |
| chr2_+_89804937 | 1.03 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chrX_+_72818003 | 0.99 |
ENSMUST00000002081.6
|
Srpk3
|
serine/arginine-rich protein specific kinase 3 |
| chr11_+_33913013 | 0.99 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr17_-_34043502 | 0.95 |
ENSMUST00000087342.13
ENSMUST00000173844.8 |
Rps28
|
ribosomal protein S28 |
| chr4_+_150572847 | 0.94 |
ENSMUST00000105680.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr1_+_163875783 | 0.91 |
ENSMUST00000027874.6
|
Sele
|
selectin, endothelial cell |
| chr1_-_134006847 | 0.89 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr13_-_93774469 | 0.88 |
ENSMUST00000099309.6
|
Bhmt
|
betaine-homocysteine methyltransferase |
| chr10_+_79540309 | 0.80 |
ENSMUST00000179781.8
ENSMUST00000067036.12 ENSMUST00000178383.8 |
Bsg
|
basigin |
| chr7_-_140846328 | 0.78 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chr19_+_8866217 | 0.77 |
ENSMUST00000096251.10
ENSMUST00000191089.2 |
1810009A15Rik
Gm49416
|
RIKEN cDNA 1810009A15 gene predicted gene, 49416 |
| chr9_+_44684206 | 0.76 |
ENSMUST00000002099.11
|
Ift46
|
intraflagellar transport 46 |
| chr11_+_62441992 | 0.75 |
ENSMUST00000019649.4
|
Ubb
|
ubiquitin B |
| chr17_-_57535003 | 0.75 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr7_+_119773070 | 0.74 |
ENSMUST00000033201.7
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr7_+_102790086 | 0.73 |
ENSMUST00000118682.3
|
Olfr588-ps1
|
olfactory receptor 588, pseudogene 1 |
| chr14_-_47426863 | 0.73 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr8_-_84467798 | 0.72 |
ENSMUST00000075843.13
ENSMUST00000109802.3 ENSMUST00000166939.8 ENSMUST00000002964.14 |
Adgre5
|
adhesion G protein-coupled receptor E5 |
| chr9_+_57818384 | 0.67 |
ENSMUST00000217129.2
|
Ubl7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chr7_+_28510350 | 0.66 |
ENSMUST00000174477.8
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr11_+_94827050 | 0.63 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr1_+_93163553 | 0.62 |
ENSMUST00000062202.14
|
Sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr15_+_98350469 | 0.62 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr2_+_119449192 | 0.61 |
ENSMUST00000028771.8
|
Nusap1
|
nucleolar and spindle associated protein 1 |
| chr13_+_17869727 | 0.61 |
ENSMUST00000221480.2
|
Mplkip
|
M-phase specific PLK1 intereacting protein |
| chr7_+_141276575 | 0.59 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr13_+_30520416 | 0.57 |
ENSMUST00000222503.2
ENSMUST00000222370.2 ENSMUST00000066412.8 ENSMUST00000223201.2 |
Agtr1a
|
angiotensin II receptor, type 1a |
| chr2_-_88590348 | 0.56 |
ENSMUST00000131038.4
ENSMUST00000213138.3 |
Olfr1199
|
olfactory receptor 1199 |
| chr10_+_73657689 | 0.56 |
ENSMUST00000064562.14
ENSMUST00000193174.6 ENSMUST00000105426.10 ENSMUST00000129404.9 ENSMUST00000131321.9 ENSMUST00000126920.9 ENSMUST00000147189.9 ENSMUST00000105424.10 ENSMUST00000092420.13 ENSMUST00000105429.10 ENSMUST00000193361.6 ENSMUST00000131724.9 ENSMUST00000152655.9 ENSMUST00000151116.9 ENSMUST00000155701.9 ENSMUST00000152819.9 ENSMUST00000125517.9 ENSMUST00000124046.8 ENSMUST00000146682.8 ENSMUST00000177107.8 ENSMUST00000149977.9 ENSMUST00000191854.6 |
Pcdh15
|
protocadherin 15 |
| chr12_+_33479604 | 0.54 |
ENSMUST00000020877.9
|
Polr1f
|
RNA polymerase I subunit F |
| chr16_+_48662894 | 0.54 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr3_+_142470806 | 0.54 |
ENSMUST00000029938.10
|
Gtf2b
|
general transcription factor IIB |
| chr7_+_28510310 | 0.52 |
ENSMUST00000038572.15
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr6_-_87958611 | 0.51 |
ENSMUST00000056403.7
|
H1f10
|
H1.10 linker histone |
| chr11_-_50101592 | 0.51 |
ENSMUST00000143379.2
ENSMUST00000015981.12 ENSMUST00000102774.11 |
Sqstm1
|
sequestosome 1 |
| chr8_+_34582184 | 0.51 |
ENSMUST00000095345.5
|
Mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr9_-_81515865 | 0.51 |
ENSMUST00000183482.2
|
Htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr4_+_118965992 | 0.48 |
ENSMUST00000134105.8
ENSMUST00000144329.8 ENSMUST00000208090.2 |
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr12_-_40249489 | 0.48 |
ENSMUST00000220951.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr13_+_23879775 | 0.48 |
ENSMUST00000041052.5
|
H1f6
|
H1.6 linker histone, cluster member |
| chr4_-_41098174 | 0.44 |
ENSMUST00000055327.8
|
Aqp3
|
aquaporin 3 |
| chr2_+_87854404 | 0.44 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr7_-_109380745 | 0.43 |
ENSMUST00000207400.2
ENSMUST00000033331.7 |
Nrip3
|
nuclear receptor interacting protein 3 |
| chr19_+_34560922 | 0.43 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr4_+_63280675 | 0.43 |
ENSMUST00000075341.4
|
Orm2
|
orosomucoid 2 |
| chr1_+_153750081 | 0.43 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chr17_+_34043536 | 0.43 |
ENSMUST00000048249.8
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chr11_-_73800125 | 0.42 |
ENSMUST00000215690.2
|
Olfr395
|
olfactory receptor 395 |
| chr15_+_32244947 | 0.42 |
ENSMUST00000067458.7
|
Sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr3_-_20329823 | 0.41 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr2_+_13578738 | 0.41 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr2_-_166904902 | 0.41 |
ENSMUST00000048988.14
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr2_-_89671899 | 0.41 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr7_-_5496955 | 0.41 |
ENSMUST00000233742.2
ENSMUST00000233941.2 ENSMUST00000233390.2 ENSMUST00000233239.2 ENSMUST00000232789.2 ENSMUST00000086297.8 |
Vmn2r28
|
vomeronasal 2, receptor 28 |
| chr7_-_141241632 | 0.40 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr11_-_74228504 | 0.40 |
ENSMUST00000213831.2
|
Olfr410
|
olfactory receptor 410 |
| chr2_+_36674288 | 0.37 |
ENSMUST00000213498.2
ENSMUST00000214909.3 ENSMUST00000215199.2 |
Olfr348
|
olfactory receptor 348 |
| chr7_-_44393654 | 0.36 |
ENSMUST00000149011.9
|
Zfp473
|
zinc finger protein 473 |
| chr18_-_15536747 | 0.36 |
ENSMUST00000079081.8
|
Aqp4
|
aquaporin 4 |
| chr7_+_18721325 | 0.36 |
ENSMUST00000063563.9
|
Nanos2
|
nanos C2HC-type zinc finger 2 |
| chr18_+_34757666 | 0.33 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr6_-_41012435 | 0.33 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr6_+_90179768 | 0.32 |
ENSMUST00000078371.6
|
V1ra8
|
vomeronasal 1 receptor, A8 |
| chr11_+_99755302 | 0.31 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr6_-_87649124 | 0.31 |
ENSMUST00000065997.5
|
Aplf
|
aprataxin and PNKP like factor |
| chr11_-_53313950 | 0.30 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr1_-_166475262 | 0.30 |
ENSMUST00000046662.8
|
Gm4847
|
predicted gene 4847 |
| chr17_+_7592045 | 0.29 |
ENSMUST00000095726.11
ENSMUST00000128533.8 ENSMUST00000129709.8 ENSMUST00000147803.8 ENSMUST00000140192.8 ENSMUST00000138222.8 ENSMUST00000144861.2 |
Tcp10a
|
t-complex protein 10a |
| chr11_-_77616103 | 0.28 |
ENSMUST00000078623.5
|
Cryba1
|
crystallin, beta A1 |
| chr10_+_77697785 | 0.27 |
ENSMUST00000218443.2
|
Gm3285
|
predicted gene 3285 |
| chr13_+_113931038 | 0.27 |
ENSMUST00000091201.7
|
Arl15
|
ADP-ribosylation factor-like 15 |
| chr11_-_74228857 | 0.27 |
ENSMUST00000214490.2
|
Olfr410
|
olfactory receptor 410 |
| chr5_-_8672951 | 0.26 |
ENSMUST00000047485.15
ENSMUST00000115378.2 |
Rundc3b
|
RUN domain containing 3B |
| chr9_-_70564403 | 0.25 |
ENSMUST00000213380.2
ENSMUST00000049031.6 |
Mindy2
|
MINDY lysine 48 deubiquitinase 2 |
| chr2_+_158099986 | 0.24 |
ENSMUST00000109499.8
ENSMUST00000109500.8 ENSMUST00000065039.3 |
Bpi
|
bactericidal permeablility increasing protein |
| chr1_+_171052623 | 0.24 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr6_+_122603369 | 0.24 |
ENSMUST00000049644.9
|
Dppa3
|
developmental pluripotency-associated 3 |
| chr15_+_10952418 | 0.23 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr17_-_56424577 | 0.22 |
ENSMUST00000019808.12
|
Plin5
|
perilipin 5 |
| chr3_+_137329433 | 0.22 |
ENSMUST00000053855.8
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr12_+_102521225 | 0.22 |
ENSMUST00000021610.7
|
Chga
|
chromogranin A |
| chr6_-_57802040 | 0.22 |
ENSMUST00000127485.8
|
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr11_-_58521327 | 0.22 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr4_+_150939521 | 0.21 |
ENSMUST00000030811.2
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr7_+_31075005 | 0.21 |
ENSMUST00000179481.2
|
Scgb1b3
|
secretoglobin, family 1B, member 3 |
| chr13_+_42454922 | 0.20 |
ENSMUST00000021796.9
|
Edn1
|
endothelin 1 |
| chr6_-_132291714 | 0.20 |
ENSMUST00000048686.9
|
Prpmp5
|
proline-rich protein MP5 |
| chr6_+_121323577 | 0.20 |
ENSMUST00000032200.16
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr6_+_42263609 | 0.19 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr7_-_45109075 | 0.19 |
ENSMUST00000210864.2
|
Ftl1
|
ferritin light polypeptide 1 |
| chr2_+_123931879 | 0.19 |
ENSMUST00000103241.8
|
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr7_+_45070244 | 0.19 |
ENSMUST00000072453.8
|
Lhb
|
luteinizing hormone beta |
| chr17_-_67354971 | 0.18 |
ENSMUST00000224862.2
|
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chr18_-_78166539 | 0.18 |
ENSMUST00000160292.8
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr15_-_101694895 | 0.18 |
ENSMUST00000071104.6
|
Krt72
|
keratin 72 |
| chr8_-_110894836 | 0.18 |
ENSMUST00000003754.8
ENSMUST00000212297.2 |
Calb2
|
calbindin 2 |
| chr11_+_106256298 | 0.18 |
ENSMUST00000009354.10
|
Prr29
|
proline rich 29 |
| chr2_-_35994072 | 0.18 |
ENSMUST00000112961.10
ENSMUST00000112966.10 |
Lhx6
|
LIM homeobox protein 6 |
| chrX_+_72271878 | 0.17 |
ENSMUST00000105111.4
|
F8a
|
factor 8-associated gene A |
| chr7_+_45175754 | 0.17 |
ENSMUST00000211227.2
ENSMUST00000051810.15 |
Plekha4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr14_+_67982630 | 0.17 |
ENSMUST00000223929.2
ENSMUST00000111095.4 |
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr4_+_128653051 | 0.17 |
ENSMUST00000030585.8
|
A3galt2
|
alpha 1,3-galactosyltransferase 2 (isoglobotriaosylceramide synthase) |
| chr2_-_6217844 | 0.16 |
ENSMUST00000042658.5
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr18_-_35836161 | 0.16 |
ENSMUST00000025208.7
|
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr6_+_70640233 | 0.16 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr6_-_130363837 | 0.16 |
ENSMUST00000032288.6
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr10_-_79875479 | 0.16 |
ENSMUST00000004786.10
|
Polr2e
|
polymerase (RNA) II (DNA directed) polypeptide E |
| chr2_-_31700592 | 0.15 |
ENSMUST00000057407.3
|
Qrfp
|
pyroglutamylated RFamide peptide |
| chr17_+_34174782 | 0.15 |
ENSMUST00000025178.17
|
Vps52
|
VPS52 GARP complex subunit |
| chr5_+_88073438 | 0.15 |
ENSMUST00000001667.13
ENSMUST00000113267.8 |
Csn3
|
casein kappa |
| chr3_-_75389047 | 0.15 |
ENSMUST00000193989.4
ENSMUST00000203169.3 |
Wdr49
|
WD repeat domain 49 |
| chr2_-_101459303 | 0.15 |
ENSMUST00000111231.10
|
Iftap
|
intraflagellar transport associated protein |
| chr1_+_127657142 | 0.15 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chrM_+_7779 | 0.14 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr2_+_154149122 | 0.14 |
ENSMUST00000088921.6
|
Bpifb9b
|
BPI fold containing family B, member 9B |
| chr2_+_13579092 | 0.14 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr6_-_113577606 | 0.14 |
ENSMUST00000035870.5
|
Fancd2os
|
Fancd2 opposite strand |
| chr4_-_63663344 | 0.13 |
ENSMUST00000239480.2
ENSMUST00000062246.8 |
Tnfsf15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr8_+_124138163 | 0.13 |
ENSMUST00000071134.4
ENSMUST00000212743.2 |
Tubb3
|
tubulin, beta 3 class III |
| chr4_-_40269778 | 0.13 |
ENSMUST00000042575.7
|
Topors
|
topoisomerase I binding, arginine/serine-rich |
| chr9_+_108385247 | 0.13 |
ENSMUST00000207810.2
ENSMUST00000207862.2 ENSMUST00000208581.2 ENSMUST00000134939.9 ENSMUST00000207713.2 |
Qars
|
glutaminyl-tRNA synthetase |
| chr17_-_56424265 | 0.13 |
ENSMUST00000113072.3
|
Plin5
|
perilipin 5 |
| chr5_+_48140480 | 0.13 |
ENSMUST00000173107.8
ENSMUST00000174313.8 ENSMUST00000174421.8 ENSMUST00000170109.9 |
Slit2
|
slit guidance ligand 2 |
| chr19_+_16416664 | 0.13 |
ENSMUST00000237350.2
|
Gna14
|
guanine nucleotide binding protein, alpha 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.7 | 5.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.5 | 8.8 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.5 | 5.5 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.4 | 3.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.3 | 1.7 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.3 | 1.0 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.3 | 2.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 2.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 1.2 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.2 | 0.9 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 0.6 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 0.6 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.2 | 0.8 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 2.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.7 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.2 | 1.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 0.8 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.7 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.9 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.1 | 0.8 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.5 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 4.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.7 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 1.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 1.1 | GO:0090241 | hypermethylation of CpG island(GO:0044027) negative regulation of histone H4 acetylation(GO:0090241) apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.6 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.5 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 1.3 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.4 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.1 | 0.5 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.2 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 1.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.2 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.1 | 0.2 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 3.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.2 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.2 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.4 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 1.8 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.9 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.9 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 1.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0060450 | positive regulation of hindgut contraction(GO:0060450) |
| 0.0 | 0.8 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.0 | GO:0003097 | renal water transport(GO:0003097) glycerol transport(GO:0015793) |
| 0.0 | 0.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0044704 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 2.8 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.4 | 1.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 0.6 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.2 | 2.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 0.5 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 3.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.7 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 1.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 7.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 2.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.4 | 22.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 1.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.4 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.4 | 1.2 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.4 | 3.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 2.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.3 | 1.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 5.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 7.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 3.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.6 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 2.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 1.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 1.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 2.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.3 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.1 | 0.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.2 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.1 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.0 | 1.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 1.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 4.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 5.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.1 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |