Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
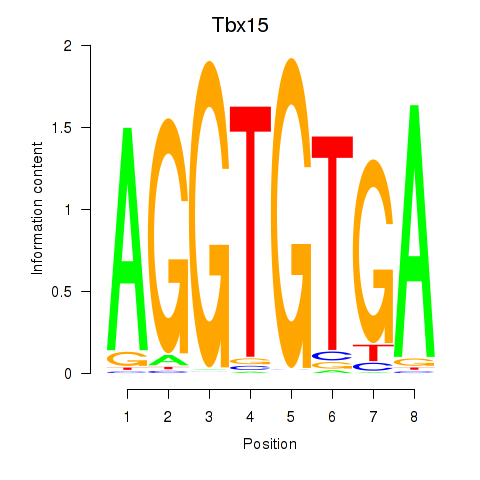
Results for Tbx15
Z-value: 0.88
Transcription factors associated with Tbx15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx15
|
ENSMUSG00000027868.12 | T-box 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx15 | mm39_v1_chr3_+_99203818_99203829 | -0.58 | 3.0e-01 | Click! |
Activity profile of Tbx15 motif
Sorted Z-values of Tbx15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_116693849 | 1.25 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr13_+_23737436 | 0.99 |
ENSMUST00000045301.9
|
H1f3
|
H1.3 linker histone, cluster member |
| chr19_+_8975249 | 0.77 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr7_+_79460475 | 0.72 |
ENSMUST00000107394.3
|
Mesp2
|
mesoderm posterior 2 |
| chr11_+_98828495 | 0.70 |
ENSMUST00000107475.9
ENSMUST00000068133.10 |
Rara
|
retinoic acid receptor, alpha |
| chr4_-_150736554 | 0.57 |
ENSMUST00000117997.2
ENSMUST00000037827.10 |
Slc45a1
|
solute carrier family 45, member 1 |
| chr6_-_28831746 | 0.56 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr12_+_33364288 | 0.54 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr6_-_97408367 | 0.49 |
ENSMUST00000124050.3
|
Frmd4b
|
FERM domain containing 4B |
| chr6_+_122803624 | 0.48 |
ENSMUST00000203075.2
|
Foxj2
|
forkhead box J2 |
| chr7_-_126224848 | 0.47 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chrM_+_9870 | 0.44 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr2_+_11647610 | 0.43 |
ENSMUST00000028111.6
|
Il2ra
|
interleukin 2 receptor, alpha chain |
| chr4_+_125384481 | 0.41 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr9_+_46910039 | 0.40 |
ENSMUST00000178065.3
|
Gm4791
|
predicted gene 4791 |
| chrX_+_41239872 | 0.39 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2 |
| chr1_-_105284383 | 0.39 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr8_+_85449632 | 0.39 |
ENSMUST00000098571.5
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr16_-_23807602 | 0.38 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr1_-_105284407 | 0.38 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr9_-_49710058 | 0.36 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr5_-_84565218 | 0.36 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr2_+_71811526 | 0.36 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chrX_+_158623460 | 0.33 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_-_86574586 | 0.31 |
ENSMUST00000018315.10
|
Vmp1
|
vacuole membrane protein 1 |
| chr1_+_17672117 | 0.31 |
ENSMUST00000088476.4
|
Pi15
|
peptidase inhibitor 15 |
| chr15_-_82783978 | 0.31 |
ENSMUST00000230403.2
|
Tcf20
|
transcription factor 20 |
| chr8_-_73229056 | 0.30 |
ENSMUST00000212991.2
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr9_-_49710190 | 0.30 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr3_-_142101339 | 0.30 |
ENSMUST00000198381.5
ENSMUST00000090134.12 ENSMUST00000196908.5 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr7_+_28834350 | 0.29 |
ENSMUST00000159975.8
ENSMUST00000094617.11 ENSMUST00000032811.12 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr7_+_44146029 | 0.29 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr11_-_23720953 | 0.29 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr7_+_5023552 | 0.29 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
| chr13_+_99321241 | 0.29 |
ENSMUST00000056558.11
|
Zfp366
|
zinc finger protein 366 |
| chr19_-_13126896 | 0.28 |
ENSMUST00000213493.2
|
Olfr1459
|
olfactory receptor 1459 |
| chr7_-_28078671 | 0.28 |
ENSMUST00000209061.2
|
Zfp36
|
zinc finger protein 36 |
| chr17_+_35354655 | 0.27 |
ENSMUST00000174478.8
ENSMUST00000174281.9 ENSMUST00000173550.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr16_+_62635039 | 0.27 |
ENSMUST00000055557.6
|
Stx19
|
syntaxin 19 |
| chr8_+_121262528 | 0.26 |
ENSMUST00000120493.8
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr8_+_84874654 | 0.26 |
ENSMUST00000143833.8
ENSMUST00000118856.8 |
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr15_-_76004395 | 0.26 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr11_-_103247150 | 0.25 |
ENSMUST00000136491.3
ENSMUST00000107023.3 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr9_+_43655230 | 0.25 |
ENSMUST00000034510.9
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr2_-_180844582 | 0.25 |
ENSMUST00000016511.6
|
Ptk6
|
PTK6 protein tyrosine kinase 6 |
| chr2_-_172782089 | 0.25 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
| chr14_-_55909314 | 0.24 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr4_+_84802592 | 0.24 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr9_-_43151179 | 0.24 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr9_-_57347366 | 0.24 |
ENSMUST00000214144.2
ENSMUST00000085709.6 ENSMUST00000214624.2 ENSMUST00000215883.2 ENSMUST00000214339.2 ENSMUST00000215299.2 ENSMUST00000214166.2 ENSMUST00000214065.2 |
Ppcdc
|
phosphopantothenoylcysteine decarboxylase |
| chr19_+_53186430 | 0.23 |
ENSMUST00000237099.2
|
Add3
|
adducin 3 (gamma) |
| chr14_-_30075424 | 0.23 |
ENSMUST00000224198.3
ENSMUST00000238675.2 ENSMUST00000112249.10 ENSMUST00000224785.3 |
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr18_-_60724855 | 0.23 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr4_+_62587101 | 0.23 |
ENSMUST00000124082.2
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr6_-_122587005 | 0.22 |
ENSMUST00000032211.5
|
Gdf3
|
growth differentiation factor 3 |
| chr6_-_124894902 | 0.22 |
ENSMUST00000032216.7
|
Ptms
|
parathymosin |
| chr11_-_70578775 | 0.22 |
ENSMUST00000036299.14
ENSMUST00000119120.2 ENSMUST00000100933.10 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr5_-_137016355 | 0.22 |
ENSMUST00000137272.2
ENSMUST00000111090.9 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr7_+_4122523 | 0.22 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr11_-_86435579 | 0.21 |
ENSMUST00000138810.3
ENSMUST00000058286.9 ENSMUST00000154617.8 |
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1 |
| chr7_+_127070615 | 0.21 |
ENSMUST00000033095.10
|
Prr14
|
proline rich 14 |
| chr4_+_42950367 | 0.21 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr16_+_44687460 | 0.21 |
ENSMUST00000102805.4
|
Cd200r2
|
Cd200 receptor 2 |
| chr2_+_68691902 | 0.21 |
ENSMUST00000176018.2
|
Cers6
|
ceramide synthase 6 |
| chrX_-_107877909 | 0.21 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chrX_-_161612373 | 0.21 |
ENSMUST00000041370.11
ENSMUST00000112316.9 ENSMUST00000112315.2 |
Txlng
|
taxilin gamma |
| chr11_+_53410552 | 0.20 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr18_+_82932747 | 0.20 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr2_+_136733421 | 0.20 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr10_+_56253418 | 0.20 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr8_-_32408864 | 0.20 |
ENSMUST00000073884.7
ENSMUST00000238812.2 |
Nrg1
|
neuregulin 1 |
| chr5_+_143608194 | 0.20 |
ENSMUST00000116456.10
|
Cyth3
|
cytohesin 3 |
| chr3_-_96833336 | 0.20 |
ENSMUST00000062944.7
|
Gja8
|
gap junction protein, alpha 8 |
| chr3_-_95041246 | 0.20 |
ENSMUST00000172572.9
ENSMUST00000173462.3 |
Scnm1
|
sodium channel modifier 1 |
| chr9_-_110474398 | 0.20 |
ENSMUST00000149089.2
|
Nbeal2
|
neurobeachin-like 2 |
| chr14_-_55909527 | 0.20 |
ENSMUST00000010520.10
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr8_-_84874468 | 0.20 |
ENSMUST00000117424.9
ENSMUST00000040383.9 |
Cc2d1a
|
coiled-coil and C2 domain containing 1A |
| chr12_-_35584968 | 0.20 |
ENSMUST00000116436.9
|
Ahr
|
aryl-hydrocarbon receptor |
| chr8_+_84874881 | 0.20 |
ENSMUST00000093375.5
|
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr19_+_4147391 | 0.19 |
ENSMUST00000174514.2
ENSMUST00000174149.8 |
Cdk2ap2
|
CDK2-associated protein 2 |
| chr11_-_70578744 | 0.19 |
ENSMUST00000108545.9
ENSMUST00000120261.8 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr1_+_156386327 | 0.19 |
ENSMUST00000173929.8
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr9_-_44632680 | 0.19 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr18_+_36693024 | 0.19 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr19_-_37184692 | 0.18 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr19_-_40260060 | 0.18 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr11_+_69804714 | 0.18 |
ENSMUST00000072581.9
ENSMUST00000116358.8 |
Gps2
|
G protein pathway suppressor 2 |
| chr13_+_56757389 | 0.18 |
ENSMUST00000045173.10
|
Tgfbi
|
transforming growth factor, beta induced |
| chr15_-_79281056 | 0.18 |
ENSMUST00000228472.2
|
Tmem184b
|
transmembrane protein 184b |
| chr2_-_44817218 | 0.17 |
ENSMUST00000100127.9
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr2_-_91013362 | 0.17 |
ENSMUST00000066420.12
|
Madd
|
MAP-kinase activating death domain |
| chr13_+_81034214 | 0.17 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr5_-_114131934 | 0.17 |
ENSMUST00000159592.8
|
Ssh1
|
slingshot protein phosphatase 1 |
| chr8_-_32408380 | 0.17 |
ENSMUST00000208497.3
ENSMUST00000207584.3 |
Nrg1
|
neuregulin 1 |
| chr7_+_126528016 | 0.17 |
ENSMUST00000032924.6
|
Kctd13
|
potassium channel tetramerisation domain containing 13 |
| chr19_+_8817883 | 0.17 |
ENSMUST00000086058.13
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr16_+_57173456 | 0.16 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr6_+_48624295 | 0.16 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
| chr4_-_127864744 | 0.16 |
ENSMUST00000030614.3
|
CK137956
|
cDNA sequence CK137956 |
| chr2_+_54326329 | 0.16 |
ENSMUST00000112636.8
ENSMUST00000112635.8 ENSMUST00000112634.8 |
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr4_-_129334593 | 0.16 |
ENSMUST00000053042.6
ENSMUST00000106046.8 |
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr7_-_78228116 | 0.16 |
ENSMUST00000206268.2
ENSMUST00000039431.14 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr16_+_84631789 | 0.16 |
ENSMUST00000114184.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chrX_+_167819606 | 0.16 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr11_+_69231589 | 0.16 |
ENSMUST00000218008.2
ENSMUST00000151617.3 |
Rnf227
|
ring finger protein 227 |
| chr7_-_28001624 | 0.16 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr3_-_142101418 | 0.15 |
ENSMUST00000029941.16
ENSMUST00000058626.9 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr9_+_102885156 | 0.15 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr17_-_57385490 | 0.15 |
ENSMUST00000011623.9
|
Dennd1c
|
DENN/MADD domain containing 1C |
| chr7_+_107194446 | 0.15 |
ENSMUST00000040056.15
ENSMUST00000208956.2 |
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr3_+_95041399 | 0.15 |
ENSMUST00000066386.6
|
Lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr11_-_98329782 | 0.15 |
ENSMUST00000002655.8
|
Mien1
|
migration and invasion enhancer 1 |
| chr5_-_116162415 | 0.15 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr4_-_41048124 | 0.15 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr4_+_134879807 | 0.15 |
ENSMUST00000119564.2
|
Runx3
|
runt related transcription factor 3 |
| chr9_-_108991088 | 0.14 |
ENSMUST00000199540.2
ENSMUST00000198076.5 ENSMUST00000054925.13 |
Fbxw21
|
F-box and WD-40 domain protein 21 |
| chr10_+_111808569 | 0.14 |
ENSMUST00000163048.8
ENSMUST00000174653.2 |
Krr1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr3_-_89294430 | 0.14 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr8_-_41087793 | 0.14 |
ENSMUST00000173957.2
ENSMUST00000048898.17 ENSMUST00000174205.8 |
Mtmr7
|
myotubularin related protein 7 |
| chr1_+_132119169 | 0.13 |
ENSMUST00000188169.7
ENSMUST00000112357.9 ENSMUST00000188175.2 |
Lemd1
Gm29695
|
LEM domain containing 1 predicted gene, 29695 |
| chr7_+_46045862 | 0.13 |
ENSMUST00000025202.8
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr1_-_87322443 | 0.13 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr11_-_120622770 | 0.13 |
ENSMUST00000154565.2
ENSMUST00000026148.9 |
Cbr2
|
carbonyl reductase 2 |
| chr17_+_35354430 | 0.13 |
ENSMUST00000173535.8
ENSMUST00000173952.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr6_-_28261881 | 0.13 |
ENSMUST00000115320.8
ENSMUST00000123098.8 ENSMUST00000115321.9 ENSMUST00000155494.2 |
Zfp800
|
zinc finger protein 800 |
| chr15_+_100513230 | 0.13 |
ENSMUST00000000356.10
|
Dazap2
|
DAZ associated protein 2 |
| chr1_+_173093568 | 0.12 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr1_-_192880260 | 0.12 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chrX_+_150799414 | 0.12 |
ENSMUST00000045312.6
|
Smc1a
|
structural maintenance of chromosomes 1A |
| chr2_+_121287444 | 0.12 |
ENSMUST00000126764.2
|
Hypk
|
huntingtin interacting protein K |
| chr18_+_82493284 | 0.12 |
ENSMUST00000047865.14
|
Mbp
|
myelin basic protein |
| chr19_+_53128901 | 0.12 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr7_-_4999099 | 0.12 |
ENSMUST00000108572.2
|
Zfp579
|
zinc finger protein 579 |
| chr15_-_82796308 | 0.12 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr9_-_37627519 | 0.12 |
ENSMUST00000215727.2
ENSMUST00000211952.3 |
Olfr160
|
olfactory receptor 160 |
| chr11_+_69657275 | 0.12 |
ENSMUST00000132528.8
ENSMUST00000153943.2 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr2_+_119004964 | 0.11 |
ENSMUST00000239130.2
ENSMUST00000069711.3 |
Gm14137
|
predicted gene 14137 |
| chr18_-_46345661 | 0.11 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr9_-_72946980 | 0.11 |
ENSMUST00000184035.8
ENSMUST00000098566.5 |
Pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr7_+_45522196 | 0.11 |
ENSMUST00000002855.14
ENSMUST00000211716.2 |
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr2_+_68691778 | 0.11 |
ENSMUST00000028426.9
|
Cers6
|
ceramide synthase 6 |
| chr17_-_24360516 | 0.11 |
ENSMUST00000115411.8
ENSMUST00000115409.9 ENSMUST00000115407.9 ENSMUST00000102927.10 |
Pdpk1
|
3-phosphoinositide dependent protein kinase 1 |
| chr4_+_136150835 | 0.11 |
ENSMUST00000088677.5
ENSMUST00000121571.8 ENSMUST00000117699.2 |
Htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D |
| chr4_-_82803384 | 0.10 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr4_-_46404224 | 0.10 |
ENSMUST00000107764.9
|
Hemgn
|
hemogen |
| chr14_+_54032814 | 0.10 |
ENSMUST00000103671.4
|
Trav13-5
|
T cell receptor alpha variable 13-5 |
| chr9_+_27210500 | 0.10 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr18_-_55123153 | 0.10 |
ENSMUST00000064763.7
|
Zfp608
|
zinc finger protein 608 |
| chr1_-_74788013 | 0.10 |
ENSMUST00000188073.7
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr18_-_37777238 | 0.10 |
ENSMUST00000066272.6
|
Taf7
|
TATA-box binding protein associated factor 7 |
| chr2_-_91013193 | 0.10 |
ENSMUST00000111370.9
ENSMUST00000111376.8 ENSMUST00000099723.9 |
Madd
|
MAP-kinase activating death domain |
| chr15_-_11905697 | 0.10 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr10_-_34294461 | 0.10 |
ENSMUST00000213269.2
ENSMUST00000099973.4 ENSMUST00000105512.8 ENSMUST00000047885.14 |
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr16_+_33201227 | 0.10 |
ENSMUST00000232023.2
|
Zfp148
|
zinc finger protein 148 |
| chr14_+_52254341 | 0.10 |
ENSMUST00000228408.2
ENSMUST00000227295.2 |
Tmem253
|
transmembrane protein 253 |
| chr11_+_74816640 | 0.10 |
ENSMUST00000045281.13
|
Smg6
|
Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr9_+_95836839 | 0.10 |
ENSMUST00000189106.2
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr11_-_54853729 | 0.09 |
ENSMUST00000108885.8
ENSMUST00000102730.9 ENSMUST00000018482.13 ENSMUST00000108886.8 ENSMUST00000102731.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr11_+_53410697 | 0.09 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr10_-_127177729 | 0.09 |
ENSMUST00000026474.5
ENSMUST00000219671.2 |
Gli1
|
GLI-Kruppel family member GLI1 |
| chr19_-_46315543 | 0.09 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr3_+_75464837 | 0.09 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr9_+_95836869 | 0.09 |
ENSMUST00000190665.2
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr4_+_19818718 | 0.09 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr8_+_12807001 | 0.09 |
ENSMUST00000033818.10
ENSMUST00000091237.12 |
Atp11a
|
ATPase, class VI, type 11A |
| chr14_+_11307729 | 0.09 |
ENSMUST00000160956.2
ENSMUST00000160340.8 ENSMUST00000162278.8 |
Fhit
|
fragile histidine triad gene |
| chr7_-_126497421 | 0.09 |
ENSMUST00000121532.8
ENSMUST00000032926.12 |
Tmem219
|
transmembrane protein 219 |
| chr16_+_84631956 | 0.09 |
ENSMUST00000009120.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr13_-_43457626 | 0.09 |
ENSMUST00000055341.7
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr12_-_113386312 | 0.09 |
ENSMUST00000177715.8
ENSMUST00000103426.3 |
Ighm
|
immunoglobulin heavy constant mu |
| chr11_-_98040377 | 0.09 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr7_+_27151838 | 0.09 |
ENSMUST00000108357.8
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr7_+_4240697 | 0.09 |
ENSMUST00000117550.2
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr13_-_96678844 | 0.09 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr6_+_72281587 | 0.09 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr2_+_164664920 | 0.09 |
ENSMUST00000132282.2
|
Zswim1
|
zinc finger SWIM-type containing 1 |
| chr11_-_23845207 | 0.09 |
ENSMUST00000102863.3
ENSMUST00000020513.10 |
Papolg
|
poly(A) polymerase gamma |
| chr14_+_65187485 | 0.09 |
ENSMUST00000043914.8
ENSMUST00000239450.2 |
Ints9
|
integrator complex subunit 9 |
| chr2_+_27566452 | 0.09 |
ENSMUST00000129514.8
|
Rxra
|
retinoid X receptor alpha |
| chr2_+_140237229 | 0.09 |
ENSMUST00000110067.8
ENSMUST00000110063.8 ENSMUST00000110064.8 ENSMUST00000110062.8 ENSMUST00000043836.8 ENSMUST00000078027.12 |
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chrM_+_10167 | 0.09 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr10_+_75047897 | 0.09 |
ENSMUST00000218766.2
|
Specc1l
|
sperm antigen with calponin homology and coiled-coil domains 1-like |
| chr14_-_45096827 | 0.09 |
ENSMUST00000095959.2
|
Ptgdr
|
prostaglandin D receptor |
| chr19_+_46044972 | 0.08 |
ENSMUST00000111899.8
ENSMUST00000099392.10 ENSMUST00000062322.11 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr7_-_44498305 | 0.08 |
ENSMUST00000207293.2
ENSMUST00000207532.2 |
Tbc1d17
|
TBC1 domain family, member 17 |
| chr1_+_171157137 | 0.08 |
ENSMUST00000142063.8
ENSMUST00000129116.8 |
Dedd
|
death effector domain-containing |
| chr7_-_103674780 | 0.08 |
ENSMUST00000218535.2
|
Olfr640
|
olfactory receptor 640 |
| chr8_-_81466126 | 0.08 |
ENSMUST00000043359.9
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr6_+_124281607 | 0.08 |
ENSMUST00000032234.5
ENSMUST00000112541.8 |
Cd163
|
CD163 antigen |
| chr15_-_44291226 | 0.08 |
ENSMUST00000227843.2
|
Nudcd1
|
NudC domain containing 1 |
| chr18_-_12369351 | 0.08 |
ENSMUST00000025279.6
|
Npc1
|
NPC intracellular cholesterol transporter 1 |
| chr19_+_23881821 | 0.08 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr2_-_10053381 | 0.08 |
ENSMUST00000026888.11
|
Taf3
|
TATA-box binding protein associated factor 3 |
| chr2_-_57004933 | 0.08 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr6_+_61157279 | 0.08 |
ENSMUST00000126214.8
|
Ccser1
|
coiled-coil serine rich 1 |
| chr4_-_59549314 | 0.08 |
ENSMUST00000148331.9
ENSMUST00000030076.12 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr6_+_108190163 | 0.08 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr8_+_23525101 | 0.08 |
ENSMUST00000117662.8
ENSMUST00000117296.8 ENSMUST00000141784.9 |
Ank1
|
ankyrin 1, erythroid |
| chr16_+_93526987 | 0.08 |
ENSMUST00000227156.2
|
Dop1b
|
DOP1 leucine zipper like protein B |
| chr19_+_5618096 | 0.08 |
ENSMUST00000096318.4
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.7 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 0.4 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.1 | 0.3 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.3 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.8 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.2 | GO:0036118 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.2 | GO:0010645 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.2 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.2 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.4 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.2 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.5 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.4 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 0.0 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.1 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.0 | 0.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |