Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
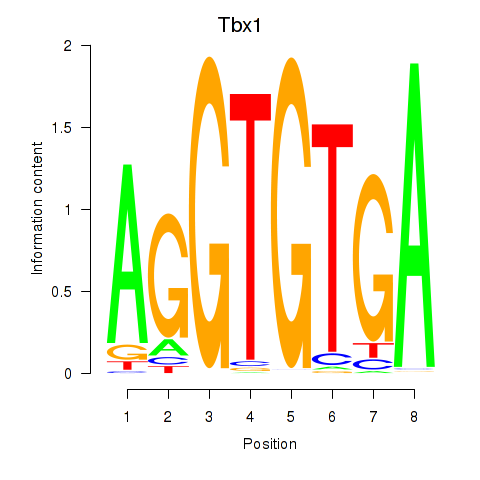
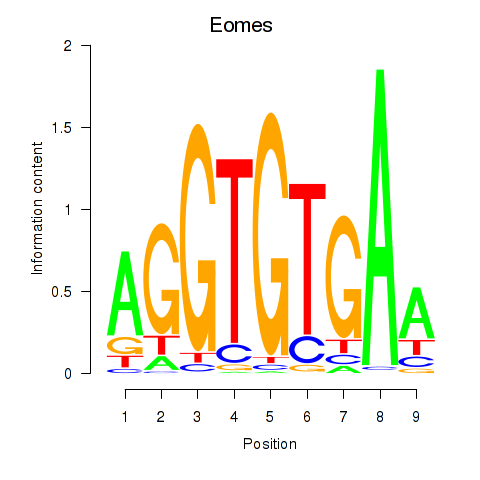
Results for Tbx1_Eomes
Z-value: 0.70
Transcription factors associated with Tbx1_Eomes
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx1
|
ENSMUSG00000009097.11 | T-box 1 |
|
Eomes
|
ENSMUSG00000032446.15 | eomesodermin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Eomes | mm39_v1_chr9_+_118307412_118307519 | 0.93 | 2.0e-02 | Click! |
| Tbx1 | mm39_v1_chr16_-_18405709_18405720 | -0.83 | 8.3e-02 | Click! |
Activity profile of Tbx1_Eomes motif
Sorted Z-values of Tbx1_Eomes motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_34218301 | 1.09 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr12_-_31684588 | 0.68 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr4_-_133746138 | 0.64 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr9_+_107468146 | 0.52 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr15_-_81244940 | 0.46 |
ENSMUST00000023040.9
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr14_+_62529924 | 0.46 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr10_+_99099084 | 0.41 |
ENSMUST00000020118.5
ENSMUST00000220291.2 |
Dusp6
|
dual specificity phosphatase 6 |
| chr10_+_26105605 | 0.40 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr12_+_3941728 | 0.40 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr11_+_23256883 | 0.37 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr2_-_30068433 | 0.37 |
ENSMUST00000100220.5
|
Spout1
|
SPOUT domain containing methyltransferase 1 |
| chr3_+_102641822 | 0.36 |
ENSMUST00000029451.12
|
Tspan2
|
tetraspanin 2 |
| chr11_+_23256909 | 0.35 |
ENSMUST00000137823.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr4_-_123644091 | 0.34 |
ENSMUST00000102636.4
|
Akirin1
|
akirin 1 |
| chr4_+_53440389 | 0.33 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr9_-_119812042 | 0.28 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr15_-_98729333 | 0.28 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr11_-_51748450 | 0.27 |
ENSMUST00000020655.14
ENSMUST00000109090.8 |
Jade2
|
jade family PHD finger 2 |
| chr11_-_117859997 | 0.27 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3 |
| chrX_+_141464393 | 0.25 |
ENSMUST00000112889.8
ENSMUST00000101198.9 ENSMUST00000112891.8 ENSMUST00000087333.9 |
Tmem164
|
transmembrane protein 164 |
| chrM_+_8603 | 0.25 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr11_+_69792642 | 0.25 |
ENSMUST00000177138.8
ENSMUST00000108617.10 ENSMUST00000177476.8 ENSMUST00000061837.11 |
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr7_+_127345909 | 0.25 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chrM_+_7758 | 0.25 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr10_-_35587888 | 0.24 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr15_+_103411689 | 0.24 |
ENSMUST00000226493.2
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr7_-_141023199 | 0.23 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr3_+_96537484 | 0.23 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr10_+_80765900 | 0.22 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr11_-_69572648 | 0.21 |
ENSMUST00000005336.9
|
Senp3
|
SUMO/sentrin specific peptidase 3 |
| chr7_-_12770556 | 0.20 |
ENSMUST00000123541.2
|
Ube2m
|
ubiquitin-conjugating enzyme E2M |
| chr11_-_69572896 | 0.20 |
ENSMUST00000066760.8
|
Senp3
|
SUMO/sentrin specific peptidase 3 |
| chr14_+_30973407 | 0.18 |
ENSMUST00000022458.11
|
Bap1
|
Brca1 associated protein 1 |
| chr17_+_47908025 | 0.18 |
ENSMUST00000183206.2
|
Ccnd3
|
cyclin D3 |
| chr11_+_98632696 | 0.17 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr7_-_45480200 | 0.17 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr8_+_124124493 | 0.16 |
ENSMUST00000212880.2
|
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr3_+_102965910 | 0.16 |
ENSMUST00000199367.5
ENSMUST00000199049.5 ENSMUST00000197678.5 |
Nras
|
neuroblastoma ras oncogene |
| chr18_-_43610829 | 0.16 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr11_+_98632953 | 0.16 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr3_+_51323383 | 0.16 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr11_+_80320558 | 0.15 |
ENSMUST00000173565.2
|
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr11_+_98632631 | 0.15 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chrM_+_7779 | 0.14 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr6_+_50087826 | 0.13 |
ENSMUST00000167628.2
|
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chrX_+_55500170 | 0.13 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr7_+_130179063 | 0.13 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr9_+_120758282 | 0.13 |
ENSMUST00000130466.8
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr11_+_102992508 | 0.13 |
ENSMUST00000107040.10
ENSMUST00000140372.8 ENSMUST00000024492.15 ENSMUST00000134884.8 |
Acbd4
|
acyl-Coenzyme A binding domain containing 4 |
| chr13_+_93440572 | 0.12 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr4_-_42168603 | 0.12 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr15_+_101982208 | 0.12 |
ENSMUST00000169681.3
ENSMUST00000229400.2 |
Eif4b
|
eukaryotic translation initiation factor 4B |
| chr4_-_42665763 | 0.12 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr4_-_132237804 | 0.12 |
ENSMUST00000030724.9
|
Sesn2
|
sestrin 2 |
| chr11_+_95304903 | 0.12 |
ENSMUST00000107724.9
ENSMUST00000150884.8 ENSMUST00000107722.8 ENSMUST00000127713.2 |
Spop
|
speckle-type BTB/POZ protein |
| chr11_-_113540867 | 0.12 |
ENSMUST00000136392.8
ENSMUST00000125890.8 ENSMUST00000146031.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr5_-_148336711 | 0.12 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr4_+_53440516 | 0.12 |
ENSMUST00000107651.9
ENSMUST00000107647.8 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr4_+_88640042 | 0.12 |
ENSMUST00000181601.2
|
Gm26566
|
predicted gene, 26566 |
| chr2_+_28403255 | 0.12 |
ENSMUST00000028170.15
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr9_-_13729961 | 0.11 |
ENSMUST00000150893.8
ENSMUST00000124883.8 |
Cep57
|
centrosomal protein 57 |
| chr6_-_48025845 | 0.11 |
ENSMUST00000095944.10
|
Zfp777
|
zinc finger protein 777 |
| chr8_+_123202935 | 0.11 |
ENSMUST00000146634.8
ENSMUST00000134127.2 |
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr8_-_46605196 | 0.11 |
ENSMUST00000110378.9
|
Snx25
|
sorting nexin 25 |
| chr16_+_35892437 | 0.10 |
ENSMUST00000163352.9
ENSMUST00000231468.2 |
Ccdc58
|
coiled-coil domain containing 58 |
| chr4_-_63779562 | 0.10 |
ENSMUST00000030047.3
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr4_+_116078830 | 0.10 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr10_-_85752765 | 0.10 |
ENSMUST00000037646.9
ENSMUST00000220032.2 |
Prdm4
|
PR domain containing 4 |
| chr5_-_148336574 | 0.10 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr9_-_35122261 | 0.10 |
ENSMUST00000043805.15
ENSMUST00000142595.8 ENSMUST00000127996.8 |
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chrX_-_100463810 | 0.10 |
ENSMUST00000118092.8
ENSMUST00000119699.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr6_+_39550827 | 0.10 |
ENSMUST00000145788.8
ENSMUST00000051249.13 |
Adck2
|
aarF domain containing kinase 2 |
| chr11_-_94398162 | 0.10 |
ENSMUST00000040692.9
|
Mycbpap
|
MYCBP associated protein |
| chr9_-_20984790 | 0.10 |
ENSMUST00000010348.7
|
Fdx2
|
ferredoxin 2 |
| chr15_+_99568208 | 0.09 |
ENSMUST00000023758.9
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr6_-_145811274 | 0.09 |
ENSMUST00000032386.11
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr1_-_167112784 | 0.09 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr6_-_92191878 | 0.09 |
ENSMUST00000014694.11
|
Rbsn
|
rabenosyn, RAB effector |
| chr10_-_128047658 | 0.09 |
ENSMUST00000061995.10
|
Spryd4
|
SPRY domain containing 4 |
| chrX_-_100463395 | 0.09 |
ENSMUST00000117901.8
ENSMUST00000120201.8 ENSMUST00000117637.8 ENSMUST00000134005.2 ENSMUST00000121520.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr3_-_94490023 | 0.09 |
ENSMUST00000029783.16
|
Snx27
|
sorting nexin family member 27 |
| chrX_+_41239548 | 0.09 |
ENSMUST00000069619.14
|
Stag2
|
stromal antigen 2 |
| chrX_-_52357897 | 0.09 |
ENSMUST00000114838.8
|
Fam122b
|
family with sequence similarity 122, member B |
| chr4_-_109522502 | 0.08 |
ENSMUST00000063531.5
|
Cdkn2c
|
cyclin dependent kinase inhibitor 2C |
| chr11_+_86574811 | 0.08 |
ENSMUST00000108022.8
ENSMUST00000108021.2 |
Ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr19_+_37425180 | 0.08 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr6_+_50087167 | 0.08 |
ENSMUST00000166318.8
ENSMUST00000036236.15 ENSMUST00000204545.3 ENSMUST00000036225.15 |
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr8_-_106660470 | 0.08 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr15_-_97665524 | 0.08 |
ENSMUST00000128775.9
ENSMUST00000134885.3 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr9_+_37119472 | 0.08 |
ENSMUST00000034632.10
|
Tmem218
|
transmembrane protein 218 |
| chr11_-_109364424 | 0.08 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr15_+_103411461 | 0.08 |
ENSMUST00000023132.5
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr18_-_36201664 | 0.08 |
ENSMUST00000239409.2
|
Nrg2
|
neuregulin 2 |
| chr12_-_56392646 | 0.08 |
ENSMUST00000021416.9
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr2_-_25162347 | 0.08 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr9_-_119151428 | 0.07 |
ENSMUST00000040853.11
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr1_-_151965876 | 0.07 |
ENSMUST00000044581.14
|
1700025G04Rik
|
RIKEN cDNA 1700025G04 gene |
| chr16_-_50151350 | 0.07 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chr14_-_54651442 | 0.07 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr15_-_82128538 | 0.07 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr11_-_109364046 | 0.07 |
ENSMUST00000070872.13
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr11_+_31823096 | 0.07 |
ENSMUST00000155278.2
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_+_6800275 | 0.07 |
ENSMUST00000043578.13
ENSMUST00000139756.8 ENSMUST00000131467.8 ENSMUST00000150761.8 ENSMUST00000151281.8 |
St18
|
suppression of tumorigenicity 18 |
| chr11_+_49737558 | 0.07 |
ENSMUST00000109179.9
ENSMUST00000178543.8 ENSMUST00000164643.8 ENSMUST00000020634.14 |
Mapk9
|
mitogen-activated protein kinase 9 |
| chr7_+_19197192 | 0.07 |
ENSMUST00000137613.9
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr8_+_123202882 | 0.07 |
ENSMUST00000116412.8
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr13_-_113317431 | 0.06 |
ENSMUST00000038212.14
|
Gzmk
|
granzyme K |
| chrX_+_139857640 | 0.06 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chrX_+_139857688 | 0.06 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr11_-_88608920 | 0.06 |
ENSMUST00000092794.12
|
Msi2
|
musashi RNA-binding protein 2 |
| chr2_+_28403084 | 0.06 |
ENSMUST00000135803.8
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr9_+_75213570 | 0.06 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr18_+_76374959 | 0.06 |
ENSMUST00000171256.8
|
Smad2
|
SMAD family member 2 |
| chr1_-_161704224 | 0.06 |
ENSMUST00000048377.11
|
Suco
|
SUN domain containing ossification factor |
| chr7_-_141023902 | 0.06 |
ENSMUST00000026580.12
|
Pidd1
|
p53 induced death domain protein 1 |
| chrM_+_9459 | 0.06 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr3_-_10273628 | 0.06 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr7_-_108529375 | 0.06 |
ENSMUST00000055745.5
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr3_-_53771185 | 0.06 |
ENSMUST00000122330.2
ENSMUST00000146598.8 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr14_-_54949596 | 0.06 |
ENSMUST00000064290.8
|
Cebpe
|
CCAAT/enhancer binding protein (C/EBP), epsilon |
| chr4_+_11191726 | 0.06 |
ENSMUST00000029866.16
ENSMUST00000108324.4 |
Ccne2
|
cyclin E2 |
| chr4_+_117706559 | 0.06 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_+_25162487 | 0.06 |
ENSMUST00000028341.11
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr1_+_163942833 | 0.06 |
ENSMUST00000162746.2
|
Selp
|
selectin, platelet |
| chr2_-_103627937 | 0.06 |
ENSMUST00000028607.13
|
Caprin1
|
cell cycle associated protein 1 |
| chr15_-_97665679 | 0.05 |
ENSMUST00000129223.9
ENSMUST00000126854.9 ENSMUST00000135080.2 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr10_+_116013256 | 0.05 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr19_-_10282218 | 0.05 |
ENSMUST00000039327.11
|
Dagla
|
diacylglycerol lipase, alpha |
| chr1_+_66507523 | 0.05 |
ENSMUST00000061620.17
ENSMUST00000212557.3 |
Unc80
|
unc-80, NALCN activator |
| chr18_+_21135079 | 0.05 |
ENSMUST00000234545.2
|
Rnf138
|
ring finger protein 138 |
| chr4_-_82423985 | 0.05 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr18_+_36414122 | 0.05 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr19_+_20579322 | 0.05 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr6_+_39550791 | 0.05 |
ENSMUST00000140364.8
|
Adck2
|
aarF domain containing kinase 2 |
| chr1_+_180553569 | 0.05 |
ENSMUST00000027780.6
|
Acbd3
|
acyl-Coenzyme A binding domain containing 3 |
| chr12_+_78908466 | 0.05 |
ENSMUST00000071230.8
|
Eif2s1
|
eukaryotic translation initiation factor 2, subunit 1 alpha |
| chr11_-_69791712 | 0.05 |
ENSMUST00000108621.9
ENSMUST00000100969.9 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr9_+_71123061 | 0.04 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr2_+_32498997 | 0.04 |
ENSMUST00000143625.2
ENSMUST00000128811.2 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr16_+_37597235 | 0.04 |
ENSMUST00000114763.3
|
Fstl1
|
follistatin-like 1 |
| chr10_-_86541349 | 0.04 |
ENSMUST00000020238.14
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chr6_+_42263609 | 0.04 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr10_+_4660119 | 0.04 |
ENSMUST00000105588.8
ENSMUST00000105589.2 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr3_-_94489855 | 0.04 |
ENSMUST00000107283.8
|
Snx27
|
sorting nexin family member 27 |
| chr10_-_23977810 | 0.04 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chrX_+_151789457 | 0.04 |
ENSMUST00000095755.4
|
Usp51
|
ubiquitin specific protease 51 |
| chr17_-_81035453 | 0.04 |
ENSMUST00000234133.2
ENSMUST00000112389.9 ENSMUST00000025089.9 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr11_+_96214078 | 0.04 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr8_+_79755194 | 0.04 |
ENSMUST00000119254.8
ENSMUST00000238669.2 |
Zfp827
|
zinc finger protein 827 |
| chr18_-_65527078 | 0.03 |
ENSMUST00000035548.16
|
Alpk2
|
alpha-kinase 2 |
| chr11_-_69791774 | 0.03 |
ENSMUST00000102580.10
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr18_+_76374651 | 0.03 |
ENSMUST00000168423.9
ENSMUST00000091831.13 |
Smad2
|
SMAD family member 2 |
| chr7_+_110628158 | 0.03 |
ENSMUST00000005749.6
|
Ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr10_+_116013122 | 0.03 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr7_-_102804952 | 0.03 |
ENSMUST00000061055.2
|
Olfr589
|
olfactory receptor 589 |
| chr5_-_137784943 | 0.03 |
ENSMUST00000132726.2
|
Mepce
|
methylphosphate capping enzyme |
| chr5_-_24806960 | 0.03 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr1_-_161616031 | 0.03 |
ENSMUST00000000834.4
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr19_-_18978463 | 0.03 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr9_+_35332837 | 0.03 |
ENSMUST00000119129.10
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr18_+_21134928 | 0.03 |
ENSMUST00000234536.2
ENSMUST00000052396.7 |
Rnf138
|
ring finger protein 138 |
| chr6_+_15196950 | 0.03 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr13_+_113346193 | 0.02 |
ENSMUST00000038144.9
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr1_+_180762587 | 0.02 |
ENSMUST00000037361.9
|
Lefty1
|
left right determination factor 1 |
| chr2_+_164810139 | 0.02 |
ENSMUST00000202479.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr10_+_118276949 | 0.02 |
ENSMUST00000068592.5
|
Ifng
|
interferon gamma |
| chr3_+_40905066 | 0.02 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr4_+_48045143 | 0.02 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chrX_+_41591410 | 0.02 |
ENSMUST00000005839.11
|
Sh2d1a
|
SH2 domain containing 1A |
| chr18_-_34505544 | 0.02 |
ENSMUST00000236887.2
|
Reep5
|
receptor accessory protein 5 |
| chr9_-_40915858 | 0.02 |
ENSMUST00000188848.8
ENSMUST00000034519.13 |
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr10_+_115653152 | 0.02 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr1_+_131898325 | 0.02 |
ENSMUST00000027695.8
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr7_-_119694400 | 0.02 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chrX_+_41591476 | 0.02 |
ENSMUST00000115070.8
ENSMUST00000153948.2 |
Sh2d1a
|
SH2 domain containing 1A |
| chr2_+_26969384 | 0.02 |
ENSMUST00000091233.7
|
Adamtsl2
|
ADAMTS-like 2 |
| chr8_+_106363141 | 0.02 |
ENSMUST00000005841.16
|
Ctcf
|
CCCTC-binding factor |
| chr7_-_107759078 | 0.02 |
ENSMUST00000208296.2
|
Olfr485
|
olfactory receptor 485 |
| chr1_+_153541412 | 0.02 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr15_+_80507671 | 0.02 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr9_-_40915895 | 0.02 |
ENSMUST00000180384.3
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr5_-_3852857 | 0.02 |
ENSMUST00000043551.11
|
Ankib1
|
ankyrin repeat and IBR domain containing 1 |
| chr17_-_71156639 | 0.02 |
ENSMUST00000134654.2
ENSMUST00000172229.8 ENSMUST00000127719.2 |
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr6_-_145811028 | 0.02 |
ENSMUST00000111703.2
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr7_+_107413787 | 0.02 |
ENSMUST00000084756.2
|
Olfr467
|
olfactory receptor 467 |
| chr2_-_147888816 | 0.01 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr7_+_108211487 | 0.01 |
ENSMUST00000077343.3
|
Olfr506
|
olfactory receptor 506 |
| chr5_-_24556602 | 0.01 |
ENSMUST00000036092.10
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr3_+_96537235 | 0.01 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr7_+_46445512 | 0.01 |
ENSMUST00000006774.11
ENSMUST00000165031.8 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr7_-_107631548 | 0.01 |
ENSMUST00000049719.4
|
Olfr478
|
olfactory receptor 478 |
| chr4_-_35845204 | 0.01 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr1_+_132243849 | 0.01 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr9_-_99592116 | 0.01 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr4_+_116078874 | 0.01 |
ENSMUST00000106490.3
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr16_-_32688640 | 0.01 |
ENSMUST00000089684.10
ENSMUST00000040986.15 ENSMUST00000115105.9 |
Rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr5_-_24534554 | 0.01 |
ENSMUST00000115098.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr13_+_96679233 | 0.01 |
ENSMUST00000077672.12
ENSMUST00000109444.3 |
Cert1
|
ceramide transporter 1 |
| chr11_-_99494134 | 0.01 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr7_-_107724503 | 0.01 |
ENSMUST00000210881.2
|
Olfr484
|
olfactory receptor 484 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx1_Eomes
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.1 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 0.5 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 | 0.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.3 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.4 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:1904499 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 | 0.4 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.3 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.0 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.0 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0071144 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.0 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.1 | 0.5 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.2 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0071820 | N-box binding(GO:0071820) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.0 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |