Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
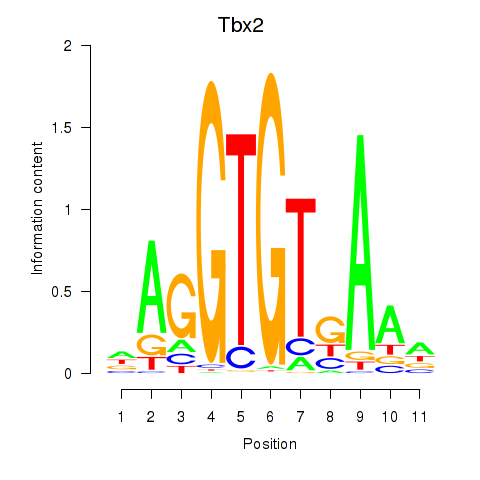
Results for Tbx2
Z-value: 0.24
Transcription factors associated with Tbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx2
|
ENSMUSG00000000093.7 | T-box 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx2 | mm39_v1_chr11_+_85723377_85723377 | -0.68 | 2.1e-01 | Click! |
Activity profile of Tbx2 motif
Sorted Z-values of Tbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_43151179 | 0.21 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr5_+_52521133 | 0.11 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr10_+_26105605 | 0.10 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr12_+_85520652 | 0.10 |
ENSMUST00000021674.7
|
Fos
|
FBJ osteosarcoma oncogene |
| chr14_+_80237691 | 0.08 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr7_+_18618605 | 0.06 |
ENSMUST00000032573.8
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr14_+_11307729 | 0.06 |
ENSMUST00000160956.2
ENSMUST00000160340.8 ENSMUST00000162278.8 |
Fhit
|
fragile histidine triad gene |
| chr4_+_94502719 | 0.06 |
ENSMUST00000107104.3
ENSMUST00000030311.11 |
Ift74
|
intraflagellar transport 74 |
| chr9_-_22043083 | 0.06 |
ENSMUST00000069330.14
ENSMUST00000217643.2 |
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr3_+_90421742 | 0.05 |
ENSMUST00000048138.8
|
S100a13
|
S100 calcium binding protein A13 |
| chr9_-_22042930 | 0.05 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr11_-_69791756 | 0.05 |
ENSMUST00000018714.13
ENSMUST00000128046.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr6_+_50087826 | 0.04 |
ENSMUST00000167628.2
|
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr9_-_49710058 | 0.04 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr15_-_82128538 | 0.04 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr6_-_28261881 | 0.04 |
ENSMUST00000115320.8
ENSMUST00000123098.8 ENSMUST00000115321.9 ENSMUST00000155494.2 |
Zfp800
|
zinc finger protein 800 |
| chr15_-_81244940 | 0.04 |
ENSMUST00000023040.9
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr2_-_103627937 | 0.04 |
ENSMUST00000028607.13
|
Caprin1
|
cell cycle associated protein 1 |
| chr3_+_95406567 | 0.04 |
ENSMUST00000015664.5
|
Ctsk
|
cathepsin K |
| chr14_-_55880708 | 0.04 |
ENSMUST00000120041.8
ENSMUST00000121937.8 ENSMUST00000133707.2 ENSMUST00000002391.15 ENSMUST00000121791.8 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr6_+_116241146 | 0.04 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr9_+_75213570 | 0.03 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr18_-_43610829 | 0.03 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr12_-_31684588 | 0.03 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr11_-_69791774 | 0.03 |
ENSMUST00000102580.10
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr1_+_88066086 | 0.03 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr11_+_98441923 | 0.03 |
ENSMUST00000081033.13
ENSMUST00000107511.8 ENSMUST00000107509.8 ENSMUST00000017339.12 |
Zpbp2
|
zona pellucida binding protein 2 |
| chr9_-_49710190 | 0.03 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr12_-_75782502 | 0.03 |
ENSMUST00000021450.6
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr7_-_30262512 | 0.03 |
ENSMUST00000207747.2
ENSMUST00000207797.2 |
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr19_+_8817883 | 0.03 |
ENSMUST00000086058.13
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr9_+_117888124 | 0.03 |
ENSMUST00000123690.2
|
Azi2
|
5-azacytidine induced gene 2 |
| chr9_+_48406706 | 0.03 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr4_+_119397710 | 0.03 |
ENSMUST00000160219.2
|
Foxj3
|
forkhead box J3 |
| chr2_+_164664920 | 0.03 |
ENSMUST00000132282.2
|
Zswim1
|
zinc finger SWIM-type containing 1 |
| chr1_-_10108325 | 0.03 |
ENSMUST00000027050.10
ENSMUST00000188619.2 |
Cops5
|
COP9 signalosome subunit 5 |
| chr11_-_69791712 | 0.03 |
ENSMUST00000108621.9
ENSMUST00000100969.9 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr3_+_96537484 | 0.03 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr5_-_137531471 | 0.03 |
ENSMUST00000143495.8
ENSMUST00000111020.8 ENSMUST00000111023.8 ENSMUST00000111038.8 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr9_-_15023396 | 0.03 |
ENSMUST00000159985.2
|
Hephl1
|
hephaestin-like 1 |
| chr3_+_96537235 | 0.03 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr1_+_171156568 | 0.03 |
ENSMUST00000111300.8
|
Dedd
|
death effector domain-containing |
| chr5_-_5564730 | 0.03 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr14_+_30973407 | 0.02 |
ENSMUST00000022458.11
|
Bap1
|
Brca1 associated protein 1 |
| chrX_-_52357897 | 0.02 |
ENSMUST00000114838.8
|
Fam122b
|
family with sequence similarity 122, member B |
| chr8_-_106223502 | 0.02 |
ENSMUST00000212303.2
|
Zdhhc1
|
zinc finger, DHHC domain containing 1 |
| chr17_+_46991972 | 0.02 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1 |
| chr7_+_19197192 | 0.02 |
ENSMUST00000137613.9
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr10_-_14420725 | 0.02 |
ENSMUST00000041168.6
ENSMUST00000238680.2 |
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr5_+_96357337 | 0.02 |
ENSMUST00000117766.8
|
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr1_+_66507523 | 0.02 |
ENSMUST00000061620.17
ENSMUST00000212557.3 |
Unc80
|
unc-80, NALCN activator |
| chr19_+_21249636 | 0.02 |
ENSMUST00000237651.2
|
Zfand5
|
zinc finger, AN1-type domain 5 |
| chr4_-_129467430 | 0.02 |
ENSMUST00000102596.8
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr4_-_120427449 | 0.02 |
ENSMUST00000030381.8
|
Ctps
|
cytidine 5'-triphosphate synthase |
| chrX_+_151789457 | 0.02 |
ENSMUST00000095755.4
|
Usp51
|
ubiquitin specific protease 51 |
| chr8_-_89362745 | 0.02 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr3_+_36606750 | 0.02 |
ENSMUST00000029269.12
ENSMUST00000136890.2 |
Exosc9
|
exosome component 9 |
| chr9_-_95727267 | 0.02 |
ENSMUST00000093800.9
|
Pls1
|
plastin 1 (I-isoform) |
| chr10_+_106306122 | 0.02 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr6_-_52135261 | 0.02 |
ENSMUST00000000964.6
ENSMUST00000120363.2 |
Hoxa1
|
homeobox A1 |
| chr1_-_171910324 | 0.02 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr9_+_72952115 | 0.02 |
ENSMUST00000184146.8
ENSMUST00000034722.5 |
Rab27a
|
RAB27A, member RAS oncogene family |
| chr14_+_52131067 | 0.02 |
ENSMUST00000047726.12
|
Slc39a2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr1_+_163942833 | 0.02 |
ENSMUST00000162746.2
|
Selp
|
selectin, platelet |
| chr4_+_53440389 | 0.02 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr5_-_3852857 | 0.01 |
ENSMUST00000043551.11
|
Ankib1
|
ankyrin repeat and IBR domain containing 1 |
| chr14_+_60940394 | 0.01 |
ENSMUST00000162131.9
|
Spata13
|
spermatogenesis associated 13 |
| chr7_-_126398165 | 0.01 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr19_-_37184692 | 0.01 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr5_-_137531413 | 0.01 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr5_-_137531463 | 0.01 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chrX_-_47123719 | 0.01 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr7_+_88079465 | 0.01 |
ENSMUST00000107256.4
|
Rab38
|
RAB38, member RAS oncogene family |
| chr6_+_42263609 | 0.01 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chrX_+_158623460 | 0.01 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr19_+_28941292 | 0.01 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr17_-_34218301 | 0.01 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr7_+_4795873 | 0.01 |
ENSMUST00000032597.12
ENSMUST00000078432.5 |
Rpl28
|
ribosomal protein L28 |
| chr9_-_48406564 | 0.01 |
ENSMUST00000213276.2
ENSMUST00000170000.4 |
Rbm7
|
RNA binding motif protein 7 |
| chr9_-_123507847 | 0.01 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr15_-_99864936 | 0.01 |
ENSMUST00000100209.6
|
Fam186a
|
family with sequence similarity 186, member A |
| chr8_-_26505605 | 0.01 |
ENSMUST00000016138.11
|
Fnta
|
farnesyltransferase, CAAX box, alpha |
| chr3_+_85878376 | 0.01 |
ENSMUST00000238443.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr19_+_37686240 | 0.01 |
ENSMUST00000025946.7
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr1_-_172156884 | 0.01 |
ENSMUST00000062387.8
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr9_-_40915858 | 0.01 |
ENSMUST00000188848.8
ENSMUST00000034519.13 |
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr9_-_20657643 | 0.01 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2 |
| chr7_+_27770655 | 0.01 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr13_+_65300040 | 0.01 |
ENSMUST00000058907.4
|
Olfr466
|
olfactory receptor 466 |
| chr19_-_10282218 | 0.01 |
ENSMUST00000039327.11
|
Dagla
|
diacylglycerol lipase, alpha |
| chr5_+_120651158 | 0.01 |
ENSMUST00000111889.2
|
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr13_-_113317431 | 0.01 |
ENSMUST00000038212.14
|
Gzmk
|
granzyme K |
| chrX_+_159865500 | 0.01 |
ENSMUST00000238603.2
|
Scml2
|
Scm polycomb group protein like 2 |
| chr5_-_5564873 | 0.01 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr2_-_113883285 | 0.01 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr2_+_52747855 | 0.01 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr9_-_40915895 | 0.01 |
ENSMUST00000180384.3
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr7_+_27307173 | 0.01 |
ENSMUST00000136962.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr1_+_131898325 | 0.01 |
ENSMUST00000027695.8
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr9_-_15834052 | 0.01 |
ENSMUST00000217187.2
|
Fat3
|
FAT atypical cadherin 3 |
| chr10_+_97528915 | 0.01 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr4_+_116078830 | 0.01 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr6_-_141801897 | 0.01 |
ENSMUST00000165990.8
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr6_-_94677118 | 0.01 |
ENSMUST00000101126.3
ENSMUST00000032105.11 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr11_+_98441998 | 0.01 |
ENSMUST00000107513.3
|
Zpbp2
|
zona pellucida binding protein 2 |
| chr1_-_51955126 | 0.01 |
ENSMUST00000046390.14
|
Myo1b
|
myosin IB |
| chr9_-_99592116 | 0.01 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr3_-_107851021 | 0.01 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr7_+_105053775 | 0.01 |
ENSMUST00000033187.6
ENSMUST00000210344.2 |
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr3_+_52175757 | 0.01 |
ENSMUST00000053764.7
|
Foxo1
|
forkhead box O1 |
| chr13_+_93444514 | 0.01 |
ENSMUST00000079086.8
|
Homer1
|
homer scaffolding protein 1 |
| chr1_+_93301596 | 0.01 |
ENSMUST00000058682.11
ENSMUST00000186641.7 |
Ano7
|
anoctamin 7 |
| chr15_-_101602734 | 0.01 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr7_-_107759078 | 0.01 |
ENSMUST00000208296.2
|
Olfr485
|
olfactory receptor 485 |
| chr15_+_6474808 | 0.01 |
ENSMUST00000022749.17
ENSMUST00000239466.2 |
C9
|
complement component 9 |
| chr13_+_48816466 | 0.01 |
ENSMUST00000021813.5
|
Barx1
|
BarH-like homeobox 1 |
| chr5_-_151113619 | 0.01 |
ENSMUST00000062015.15
ENSMUST00000110483.9 |
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr2_+_150942528 | 0.01 |
ENSMUST00000178211.2
|
Gm14147
|
predicted gene 14147 |
| chr6_-_24956296 | 0.01 |
ENSMUST00000127247.4
|
Tmem229a
|
transmembrane protein 229A |
| chr14_+_50759296 | 0.01 |
ENSMUST00000215793.2
|
Olfr743
|
olfactory receptor 743 |
| chr2_+_130038463 | 0.00 |
ENSMUST00000166774.3
|
Tmc2
|
transmembrane channel-like gene family 2 |
| chr15_-_89294434 | 0.00 |
ENSMUST00000109314.9
|
Syce3
|
synaptonemal complex central element protein 3 |
| chr6_+_15196950 | 0.00 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr2_+_151923449 | 0.00 |
ENSMUST00000064061.4
|
Scrt2
|
scratch family zinc finger 2 |
| chr10_-_23977810 | 0.00 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr10_-_128047658 | 0.00 |
ENSMUST00000061995.10
|
Spryd4
|
SPRY domain containing 4 |
| chr2_+_172821620 | 0.00 |
ENSMUST00000109125.8
ENSMUST00000109126.5 ENSMUST00000050442.15 |
Spo11
|
SPO11 initiator of meiotic double stranded breaks |
| chr6_-_141801918 | 0.00 |
ENSMUST00000163678.2
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr4_-_58785722 | 0.00 |
ENSMUST00000059608.5
|
Olfr267
|
olfactory receptor 267 |
| chr4_-_58499398 | 0.00 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr8_-_44152524 | 0.00 |
ENSMUST00000212185.2
|
Adam34
|
a disintegrin and metallopeptidase domain 34 |
| chr2_-_120946877 | 0.00 |
ENSMUST00000110675.3
|
Tgm7
|
transglutaminase 7 |
| chrX_+_100473161 | 0.00 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr4_+_116078874 | 0.00 |
ENSMUST00000106490.3
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr15_+_79113341 | 0.00 |
ENSMUST00000163571.8
|
Pick1
|
protein interacting with C kinase 1 |
| chr7_-_107424741 | 0.00 |
ENSMUST00000213252.2
|
Olfr469
|
olfactory receptor 469 |
| chr1_-_172156828 | 0.00 |
ENSMUST00000194204.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr6_+_142244145 | 0.00 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr15_-_96917804 | 0.00 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr9_-_99592058 | 0.00 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr8_+_46338557 | 0.00 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr7_+_27307422 | 0.00 |
ENSMUST00000142365.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr1_-_71454041 | 0.00 |
ENSMUST00000087268.7
|
Abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr7_+_4240697 | 0.00 |
ENSMUST00000117550.2
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr11_-_87783073 | 0.00 |
ENSMUST00000213672.2
ENSMUST00000213928.2 |
Olfr462
|
olfactory receptor 462 |
| chr10_-_23968192 | 0.00 |
ENSMUST00000092654.4
|
Taar8b
|
trace amine-associated receptor 8B |
| chrX_+_138110780 | 0.00 |
ENSMUST00000169886.8
ENSMUST00000113043.8 ENSMUST00000113045.9 |
Pwwp3b
|
PWWP domain containing 3B |
| chr3_+_95466982 | 0.00 |
ENSMUST00000090797.11
ENSMUST00000171191.6 ENSMUST00000029754.13 ENSMUST00000107154.4 |
Hormad1
|
HORMA domain containing 1 |
| chr3_-_108062172 | 0.00 |
ENSMUST00000062028.8
|
Gpr61
|
G protein-coupled receptor 61 |
| chr4_-_49549489 | 0.00 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr7_-_102804952 | 0.00 |
ENSMUST00000061055.2
|
Olfr589
|
olfactory receptor 589 |
| chr7_+_45135784 | 0.00 |
ENSMUST00000107758.10
|
Tulp2
|
tubby-like protein 2 |
| chr3_-_158267771 | 0.00 |
ENSMUST00000199890.5
ENSMUST00000238317.3 ENSMUST00000200137.5 ENSMUST00000106044.6 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr11_-_83959175 | 0.00 |
ENSMUST00000100705.11
|
Dusp14
|
dual specificity phosphatase 14 |
| chr5_-_87634665 | 0.00 |
ENSMUST00000201519.2
|
Gm43638
|
predicted gene 43638 |
| chr10_+_90412114 | 0.00 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr8_+_55024446 | 0.00 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr3_+_138047536 | 0.00 |
ENSMUST00000199673.6
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr9_+_38120348 | 0.00 |
ENSMUST00000093867.3
|
Olfr893
|
olfactory receptor 893 |
| chr6_-_141954471 | 0.00 |
ENSMUST00000111832.3
|
Gm6614
|
predicted gene 6614 |
| chr7_+_26456567 | 0.00 |
ENSMUST00000077855.8
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr1_-_156936197 | 0.00 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr7_+_88079534 | 0.00 |
ENSMUST00000208478.2
|
Rab38
|
RAB38, member RAS oncogene family |
| chrX_+_136552469 | 0.00 |
ENSMUST00000075471.4
|
Il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chrX_+_100427331 | 0.00 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr2_+_160722562 | 0.00 |
ENSMUST00000109456.9
|
Lpin3
|
lipin 3 |
| chr4_+_116078787 | 0.00 |
ENSMUST00000147292.8
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr13_+_33268095 | 0.00 |
ENSMUST00000016951.8
|
Serpinb1b
|
serine (or cysteine) peptidase inhibitor, clade B, member 1b |
| chr14_+_68321302 | 0.00 |
ENSMUST00000022639.8
|
Nefl
|
neurofilament, light polypeptide |
| chr2_+_74512638 | 0.00 |
ENSMUST00000142312.3
|
Hoxd11
|
homeobox D11 |
| chr7_-_30826184 | 0.00 |
ENSMUST00000211945.2
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr15_-_95426108 | 0.00 |
ENSMUST00000075275.3
|
Nell2
|
NEL-like 2 |
| chr1_-_87017633 | 0.00 |
ENSMUST00000027455.13
ENSMUST00000188310.2 |
Alppl2
|
alkaline phosphatase, placental-like 2 |
| chr6_-_70217956 | 0.00 |
ENSMUST00000196275.2
|
Gm43218
|
predicted gene 43218 |
| chr17_-_42922286 | 0.00 |
ENSMUST00000068355.8
|
Opn5
|
opsin 5 |
| chr9_+_38120508 | 0.00 |
ENSMUST00000212815.2
|
Olfr893
|
olfactory receptor 893 |
| chr10_+_101517556 | 0.00 |
ENSMUST00000156751.8
|
Mgat4c
|
MGAT4 family, member C |
| chrX_+_41591410 | 0.00 |
ENSMUST00000005839.11
|
Sh2d1a
|
SH2 domain containing 1A |
| chr8_+_84075066 | 0.00 |
ENSMUST00000038692.6
|
Mgat4d
|
MGAT4 family, member C |
| chr14_-_30665232 | 0.00 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr15_-_26895660 | 0.00 |
ENSMUST00000059204.11
|
Fbxl7
|
F-box and leucine-rich repeat protein 7 |
| chr6_+_42263644 | 0.00 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr1_-_190711151 | 0.00 |
ENSMUST00000047409.9
|
Vash2
|
vasohibin 2 |
| chr19_+_20470114 | 0.00 |
ENSMUST00000225313.2
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr2_-_89465327 | 0.00 |
ENSMUST00000216124.3
|
Olfr1249
|
olfactory receptor 1249 |
| chr17_-_16051295 | 0.00 |
ENSMUST00000231985.2
|
Rgmb
|
repulsive guidance molecule family member B |
| chr1_+_131526977 | 0.00 |
ENSMUST00000027690.7
|
Avpr1b
|
arginine vasopressin receptor 1B |
| chr9_-_122123475 | 0.00 |
ENSMUST00000042546.4
|
Ano10
|
anoctamin 10 |
| chr8_+_46338498 | 0.00 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr13_+_112600604 | 0.00 |
ENSMUST00000183663.8
ENSMUST00000184311.8 ENSMUST00000183886.8 |
Il6st
|
interleukin 6 signal transducer |
| chr7_-_79443536 | 0.00 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1 |
| chr14_+_53574579 | 0.00 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr19_+_20470056 | 0.00 |
ENSMUST00000225337.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr10_-_62723103 | 0.00 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr7_-_28001624 | 0.00 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr7_-_141023199 | 0.00 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr10_-_62723238 | 0.00 |
ENSMUST00000228901.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr19_-_40365318 | 0.00 |
ENSMUST00000239304.2
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr1_-_161615927 | 0.00 |
ENSMUST00000193648.2
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 0.1 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |