Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
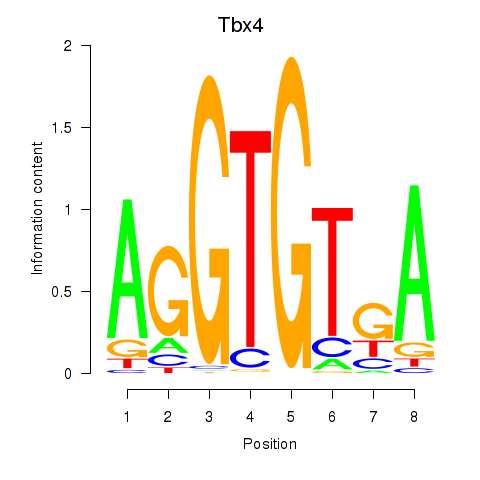
Results for Tbx4
Z-value: 0.43
Transcription factors associated with Tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx4
|
ENSMUSG00000000094.13 | T-box 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx4 | mm39_v1_chr11_+_85777185_85777248 | 0.52 | 3.7e-01 | Click! |
Activity profile of Tbx4 motif
Sorted Z-values of Tbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_116693849 | 0.35 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr7_-_103463120 | 0.28 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr2_+_32498997 | 0.22 |
ENSMUST00000143625.2
ENSMUST00000128811.2 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr7_+_79460475 | 0.19 |
ENSMUST00000107394.3
|
Mesp2
|
mesoderm posterior 2 |
| chr4_-_46404224 | 0.17 |
ENSMUST00000107764.9
|
Hemgn
|
hemogen |
| chr1_+_132243849 | 0.17 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr13_-_96678987 | 0.15 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chr6_-_28831746 | 0.13 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr18_-_46345661 | 0.13 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr19_+_5742880 | 0.13 |
ENSMUST00000235661.2
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr8_-_106660470 | 0.13 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr7_-_108529375 | 0.12 |
ENSMUST00000055745.5
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr1_-_132953068 | 0.11 |
ENSMUST00000186617.7
ENSMUST00000067429.10 ENSMUST00000067398.13 ENSMUST00000188090.7 |
Mdm4
|
transformed mouse 3T3 cell double minute 4 |
| chr9_+_102885156 | 0.11 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr4_-_19922599 | 0.11 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr11_-_98040377 | 0.11 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr4_+_125384481 | 0.10 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr11_+_121128042 | 0.10 |
ENSMUST00000103015.4
|
Narf
|
nuclear prelamin A recognition factor |
| chr9_+_108174052 | 0.10 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr18_+_5591864 | 0.10 |
ENSMUST00000025081.13
ENSMUST00000159390.8 |
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr17_-_34218301 | 0.09 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr18_-_60724855 | 0.09 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr17_+_47908025 | 0.08 |
ENSMUST00000183206.2
|
Ccnd3
|
cyclin D3 |
| chr13_-_96678844 | 0.08 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr13_+_96679110 | 0.08 |
ENSMUST00000179226.8
|
Cert1
|
ceramide transporter 1 |
| chr11_-_22236795 | 0.08 |
ENSMUST00000180360.8
ENSMUST00000109563.9 |
Ehbp1
|
EH domain binding protein 1 |
| chr11_+_69792642 | 0.08 |
ENSMUST00000177138.8
ENSMUST00000108617.10 ENSMUST00000177476.8 ENSMUST00000061837.11 |
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr2_-_91013193 | 0.08 |
ENSMUST00000111370.9
ENSMUST00000111376.8 ENSMUST00000099723.9 |
Madd
|
MAP-kinase activating death domain |
| chr19_-_32080496 | 0.07 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr13_+_108295257 | 0.07 |
ENSMUST00000120672.8
ENSMUST00000054835.15 |
Ercc8
|
excision repaiross-complementing rodent repair deficiency, complementation group 8 |
| chrX_+_99773784 | 0.07 |
ENSMUST00000113744.2
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr4_+_117706559 | 0.07 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chrX_+_99773523 | 0.07 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr12_+_3941728 | 0.07 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr3_-_94489855 | 0.06 |
ENSMUST00000107283.8
|
Snx27
|
sorting nexin family member 27 |
| chr4_-_133399909 | 0.06 |
ENSMUST00000062118.11
ENSMUST00000067902.13 |
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr1_-_57010921 | 0.06 |
ENSMUST00000114415.10
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr4_+_117706390 | 0.06 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_-_46445085 | 0.06 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr11_+_23256909 | 0.06 |
ENSMUST00000137823.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr11_-_23720953 | 0.06 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr18_+_69654572 | 0.06 |
ENSMUST00000200862.4
|
Tcf4
|
transcription factor 4 |
| chr2_-_91013362 | 0.06 |
ENSMUST00000066420.12
|
Madd
|
MAP-kinase activating death domain |
| chr15_-_76004395 | 0.05 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr8_+_79754980 | 0.05 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr10_+_86541675 | 0.05 |
ENSMUST00000075632.14
ENSMUST00000217747.2 ENSMUST00000061458.9 |
Ttc41
|
tetratricopeptide repeat domain 41 |
| chr1_+_74582044 | 0.05 |
ENSMUST00000113749.8
ENSMUST00000067916.13 ENSMUST00000113747.8 ENSMUST00000113750.8 |
Plcd4
|
phospholipase C, delta 4 |
| chr11_+_78717398 | 0.05 |
ENSMUST00000147875.9
ENSMUST00000141321.2 |
Lyrm9
|
LYR motif containing 9 |
| chr7_+_89779564 | 0.05 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr19_-_10581622 | 0.05 |
ENSMUST00000037678.7
|
Tkfc
|
triokinase, FMN cyclase |
| chr16_+_62635039 | 0.05 |
ENSMUST00000055557.6
|
Stx19
|
syntaxin 19 |
| chr2_+_28403255 | 0.04 |
ENSMUST00000028170.15
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr13_+_96679233 | 0.04 |
ENSMUST00000077672.12
ENSMUST00000109444.3 |
Cert1
|
ceramide transporter 1 |
| chr2_+_136733421 | 0.04 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr19_+_23881821 | 0.04 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr13_+_113346193 | 0.04 |
ENSMUST00000038144.9
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr12_+_119356318 | 0.04 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr4_+_11191354 | 0.04 |
ENSMUST00000170901.8
|
Ccne2
|
cyclin E2 |
| chr6_-_122587005 | 0.04 |
ENSMUST00000032211.5
|
Gdf3
|
growth differentiation factor 3 |
| chr5_-_148336574 | 0.04 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr4_+_88640042 | 0.04 |
ENSMUST00000181601.2
|
Gm26566
|
predicted gene, 26566 |
| chr3_-_94490023 | 0.04 |
ENSMUST00000029783.16
|
Snx27
|
sorting nexin family member 27 |
| chr17_-_24360516 | 0.04 |
ENSMUST00000115411.8
ENSMUST00000115409.9 ENSMUST00000115407.9 ENSMUST00000102927.10 |
Pdpk1
|
3-phosphoinositide dependent protein kinase 1 |
| chr14_-_8551429 | 0.04 |
ENSMUST00000112652.9
|
Gm11100
|
predicted gene 11100 |
| chr4_-_133746138 | 0.04 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr7_-_12770556 | 0.03 |
ENSMUST00000123541.2
|
Ube2m
|
ubiquitin-conjugating enzyme E2M |
| chr18_+_69654900 | 0.03 |
ENSMUST00000202057.4
|
Tcf4
|
transcription factor 4 |
| chr8_+_124124493 | 0.03 |
ENSMUST00000212880.2
|
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr18_+_36414122 | 0.03 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr5_-_148336711 | 0.03 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr18_+_69654992 | 0.03 |
ENSMUST00000201627.4
|
Tcf4
|
transcription factor 4 |
| chr19_-_45619559 | 0.03 |
ENSMUST00000160718.9
|
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr1_-_192880260 | 0.03 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr11_-_117859997 | 0.03 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3 |
| chr17_-_79244460 | 0.03 |
ENSMUST00000024885.10
|
Cebpz
|
CCAAT/enhancer binding protein zeta |
| chr12_-_56392646 | 0.02 |
ENSMUST00000021416.9
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr2_+_54326329 | 0.02 |
ENSMUST00000112636.8
ENSMUST00000112635.8 ENSMUST00000112634.8 |
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chrX_-_58613428 | 0.02 |
ENSMUST00000119833.8
ENSMUST00000131319.8 |
Fgf13
|
fibroblast growth factor 13 |
| chr2_-_57004933 | 0.02 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_+_23256883 | 0.02 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr9_-_102231884 | 0.02 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chr1_-_105284407 | 0.02 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr7_-_46445305 | 0.02 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr4_+_11191726 | 0.02 |
ENSMUST00000029866.16
ENSMUST00000108324.4 |
Ccne2
|
cyclin E2 |
| chr11_+_95304903 | 0.02 |
ENSMUST00000107724.9
ENSMUST00000150884.8 ENSMUST00000107722.8 ENSMUST00000127713.2 |
Spop
|
speckle-type BTB/POZ protein |
| chr15_-_78687216 | 0.02 |
ENSMUST00000164826.8
|
Card10
|
caspase recruitment domain family, member 10 |
| chr2_+_28403084 | 0.02 |
ENSMUST00000135803.8
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr13_-_89890609 | 0.02 |
ENSMUST00000109546.9
|
Vcan
|
versican |
| chr4_+_156214969 | 0.01 |
ENSMUST00000209248.2
|
Rnf223
|
ring finger 223 |
| chr11_-_86435579 | 0.01 |
ENSMUST00000138810.3
ENSMUST00000058286.9 ENSMUST00000154617.8 |
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1 |
| chr16_-_50151350 | 0.01 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chr3_+_51323383 | 0.01 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr15_+_57558048 | 0.01 |
ENSMUST00000096430.11
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr4_+_148085179 | 0.01 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr4_-_42665763 | 0.01 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr9_+_107468146 | 0.01 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr19_+_20579322 | 0.01 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr3_+_40905216 | 0.01 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr6_+_39550827 | 0.01 |
ENSMUST00000145788.8
ENSMUST00000051249.13 |
Adck2
|
aarF domain containing kinase 2 |
| chr1_+_40619215 | 0.01 |
ENSMUST00000027233.9
|
Slc9a4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4 |
| chr4_-_42168603 | 0.01 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr13_-_108295018 | 0.01 |
ENSMUST00000225830.2
ENSMUST00000223734.2 ENSMUST00000163558.3 |
Ndufaf2
|
NADH:ubiquinone oxidoreductase complex assembly factor 2 |
| chr1_-_161616031 | 0.01 |
ENSMUST00000000834.4
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr10_-_128047658 | 0.01 |
ENSMUST00000061995.10
|
Spryd4
|
SPRY domain containing 4 |
| chr1_+_153541412 | 0.01 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr13_+_81034214 | 0.01 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr15_-_11905697 | 0.01 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr19_-_42190589 | 0.01 |
ENSMUST00000018966.8
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr10_+_87697155 | 0.01 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr14_+_45588509 | 0.01 |
ENSMUST00000226873.2
|
Styx
|
serine/threonine/tyrosine interaction protein |
| chr13_-_111626562 | 0.01 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr3_-_121056944 | 0.01 |
ENSMUST00000128909.8
ENSMUST00000029777.14 |
Tlcd4
|
TLC domain containing 4 |
| chr6_-_145811274 | 0.00 |
ENSMUST00000032386.11
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr12_-_31684588 | 0.00 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chrX_+_41591355 | 0.00 |
ENSMUST00000189753.7
|
Sh2d1a
|
SH2 domain containing 1A |
| chr7_-_141023902 | 0.00 |
ENSMUST00000026580.12
|
Pidd1
|
p53 induced death domain protein 1 |
| chr18_+_65715460 | 0.00 |
ENSMUST00000169679.8
ENSMUST00000183326.2 |
Zfp532
|
zinc finger protein 532 |
| chr7_+_45890379 | 0.00 |
ENSMUST00000164538.2
|
Otog
|
otogelin |
| chr7_-_100307601 | 0.00 |
ENSMUST00000138830.2
ENSMUST00000107044.10 ENSMUST00000116287.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chrX_+_41591476 | 0.00 |
ENSMUST00000115070.8
ENSMUST00000153948.2 |
Sh2d1a
|
SH2 domain containing 1A |
| chr1_-_105284383 | 0.00 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr19_+_55882942 | 0.00 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr14_+_53574579 | 0.00 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr8_+_55024446 | 0.00 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr15_+_10952418 | 0.00 |
ENSMUST00000022853.15
ENSMUST00000110523.2 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr7_-_115423934 | 0.00 |
ENSMUST00000169129.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr4_+_53440389 | 0.00 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr15_+_98532866 | 0.00 |
ENSMUST00000230490.2
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0050354 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.0 | 0.0 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.0 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |