Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
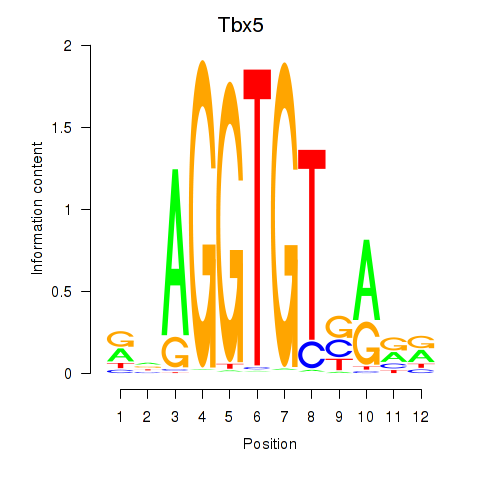
Results for Tbx5
Z-value: 0.85
Transcription factors associated with Tbx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx5
|
ENSMUSG00000018263.12 | T-box 5 |
Activity profile of Tbx5 motif
Sorted Z-values of Tbx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_79460475 | 0.88 |
ENSMUST00000107394.3
|
Mesp2
|
mesoderm posterior 2 |
| chr13_+_23737436 | 0.85 |
ENSMUST00000045301.9
|
H1f3
|
H1.3 linker histone, cluster member |
| chr7_-_6525801 | 0.80 |
ENSMUST00000213504.2
ENSMUST00000216447.2 ENSMUST00000213656.2 ENSMUST00000207820.3 |
Olfr1349
|
olfactory receptor 1349 |
| chr11_+_68986043 | 0.75 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr6_+_125026915 | 0.60 |
ENSMUST00000088294.12
ENSMUST00000032481.8 |
Acrbp
|
proacrosin binding protein |
| chr6_+_125026865 | 0.58 |
ENSMUST00000112413.8
|
Acrbp
|
proacrosin binding protein |
| chr7_+_44146029 | 0.48 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr9_+_43655230 | 0.47 |
ENSMUST00000034510.9
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr2_+_11647610 | 0.46 |
ENSMUST00000028111.6
|
Il2ra
|
interleukin 2 receptor, alpha chain |
| chr3_-_100396635 | 0.45 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr6_-_28831746 | 0.44 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr4_+_119671688 | 0.38 |
ENSMUST00000106307.9
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chrX_+_128650486 | 0.37 |
ENSMUST00000167619.9
ENSMUST00000037854.15 |
Diaph2
|
diaphanous related formin 2 |
| chr3_+_131791042 | 0.36 |
ENSMUST00000029665.7
|
Dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr14_+_50683002 | 0.36 |
ENSMUST00000214792.2
|
Olfr740
|
olfactory receptor 740 |
| chr3_-_92514799 | 0.36 |
ENSMUST00000195278.2
|
2210017I01Rik
|
RIKEN cDNA 2210017I01 gene |
| chr15_-_9748863 | 0.36 |
ENSMUST00000159368.2
ENSMUST00000159093.8 ENSMUST00000162780.8 ENSMUST00000160236.8 ENSMUST00000208854.2 ENSMUST00000041840.14 |
Spef2
|
sperm flagellar 2 |
| chr12_+_33364288 | 0.35 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr9_-_110305705 | 0.34 |
ENSMUST00000198164.5
ENSMUST00000068025.13 |
Klhl18
|
kelch-like 18 |
| chr19_+_3372296 | 0.34 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr9_+_102885156 | 0.31 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr2_-_104324035 | 0.31 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr17_+_32725420 | 0.30 |
ENSMUST00000235238.2
ENSMUST00000165999.2 |
Cyp4f17
|
cytochrome P450, family 4, subfamily f, polypeptide 17 |
| chr11_+_118319029 | 0.30 |
ENSMUST00000124861.2
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr1_-_105284407 | 0.30 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr10_-_90918291 | 0.30 |
ENSMUST00000161987.8
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr6_-_70237939 | 0.30 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr3_-_94693740 | 0.30 |
ENSMUST00000153263.9
ENSMUST00000107272.7 ENSMUST00000155485.4 |
Cgn
|
cingulin |
| chr7_+_24990596 | 0.29 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr11_+_6339061 | 0.29 |
ENSMUST00000109787.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr1_-_84262274 | 0.29 |
ENSMUST00000177458.2
ENSMUST00000168574.9 |
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr1_-_105284383 | 0.29 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr15_-_98769056 | 0.28 |
ENSMUST00000178486.9
ENSMUST00000023741.16 |
Kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr1_+_132119169 | 0.27 |
ENSMUST00000188169.7
ENSMUST00000112357.9 ENSMUST00000188175.2 |
Lemd1
Gm29695
|
LEM domain containing 1 predicted gene, 29695 |
| chr2_-_27317004 | 0.27 |
ENSMUST00000056176.8
|
Vav2
|
vav 2 oncogene |
| chr5_+_98328723 | 0.27 |
ENSMUST00000112959.4
|
Prdm8
|
PR domain containing 8 |
| chr1_-_131455039 | 0.26 |
ENSMUST00000097588.9
ENSMUST00000186543.7 |
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr12_-_35584968 | 0.26 |
ENSMUST00000116436.9
|
Ahr
|
aryl-hydrocarbon receptor |
| chr6_+_3993774 | 0.25 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr11_+_49094292 | 0.25 |
ENSMUST00000150284.8
ENSMUST00000109197.8 ENSMUST00000151228.2 |
Zfp62
|
zinc finger protein 62 |
| chr18_+_82932747 | 0.25 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr7_-_28297565 | 0.25 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr15_-_76004395 | 0.25 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr9_+_44410417 | 0.25 |
ENSMUST00000074989.7
ENSMUST00000218913.2 |
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr8_+_23525101 | 0.24 |
ENSMUST00000117662.8
ENSMUST00000117296.8 ENSMUST00000141784.9 |
Ank1
|
ankyrin 1, erythroid |
| chr2_-_91013193 | 0.24 |
ENSMUST00000111370.9
ENSMUST00000111376.8 ENSMUST00000099723.9 |
Madd
|
MAP-kinase activating death domain |
| chrX_+_128650579 | 0.23 |
ENSMUST00000113320.3
|
Diaph2
|
diaphanous related formin 2 |
| chr4_+_132291369 | 0.23 |
ENSMUST00000070690.8
|
Ptafr
|
platelet-activating factor receptor |
| chr5_-_91550853 | 0.23 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr7_+_24967094 | 0.22 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor |
| chr18_-_46345661 | 0.22 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr4_+_84802592 | 0.22 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr12_-_114252202 | 0.22 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr16_+_44687460 | 0.22 |
ENSMUST00000102805.4
|
Cd200r2
|
Cd200 receptor 2 |
| chr15_-_84804239 | 0.22 |
ENSMUST00000189185.2
|
Gm29666
|
predicted gene 29666 |
| chr5_-_142892457 | 0.22 |
ENSMUST00000167721.8
ENSMUST00000163829.2 ENSMUST00000100497.11 |
Actb
|
actin, beta |
| chr11_-_23720953 | 0.22 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr10_+_60182630 | 0.21 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr6_-_136638926 | 0.21 |
ENSMUST00000032336.7
|
Plbd1
|
phospholipase B domain containing 1 |
| chr10_+_122514669 | 0.21 |
ENSMUST00000161487.8
ENSMUST00000067918.12 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr4_-_132125510 | 0.21 |
ENSMUST00000136711.2
ENSMUST00000084249.11 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr13_+_13764982 | 0.21 |
ENSMUST00000110559.3
|
Lyst
|
lysosomal trafficking regulator |
| chr15_-_82917495 | 0.21 |
ENSMUST00000231165.2
|
Nfam1
|
Nfat activating molecule with ITAM motif 1 |
| chr4_+_83443777 | 0.20 |
ENSMUST00000053414.13
ENSMUST00000231339.2 ENSMUST00000126429.8 |
Ccdc171
|
coiled-coil domain containing 171 |
| chr11_+_6339442 | 0.19 |
ENSMUST00000109786.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr2_-_44817218 | 0.19 |
ENSMUST00000100127.9
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr6_+_35229589 | 0.19 |
ENSMUST00000152147.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr11_-_69692542 | 0.19 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chrX_+_158623460 | 0.18 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr8_+_121264161 | 0.18 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr8_+_3403415 | 0.18 |
ENSMUST00000098966.5
ENSMUST00000239102.2 |
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18 |
| chr8_+_106245368 | 0.18 |
ENSMUST00000034363.7
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr8_+_121262528 | 0.18 |
ENSMUST00000120493.8
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chrX_-_73009933 | 0.18 |
ENSMUST00000114372.3
ENSMUST00000033761.13 |
Hcfc1
|
host cell factor C1 |
| chr11_+_6339330 | 0.18 |
ENSMUST00000012612.11
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr13_+_118851214 | 0.17 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr2_-_38816229 | 0.17 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chrX_+_167819606 | 0.17 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr2_-_91013362 | 0.17 |
ENSMUST00000066420.12
|
Madd
|
MAP-kinase activating death domain |
| chr12_-_111638722 | 0.17 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr10_+_56253418 | 0.16 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr2_-_126985226 | 0.16 |
ENSMUST00000110386.2
ENSMUST00000132773.2 |
Itpripl1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1 |
| chr8_-_106016496 | 0.16 |
ENSMUST00000014981.8
|
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr19_-_46315543 | 0.16 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr6_-_146403410 | 0.16 |
ENSMUST00000053273.15
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr17_-_46857199 | 0.16 |
ENSMUST00000182485.8
ENSMUST00000066026.8 |
Cul9
|
cullin 9 |
| chr13_+_81034214 | 0.16 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr11_+_53410552 | 0.15 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr11_+_70529944 | 0.15 |
ENSMUST00000055184.7
ENSMUST00000108551.3 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr9_+_88721217 | 0.15 |
ENSMUST00000163255.9
ENSMUST00000186363.2 |
Trim43c
|
tripartite motif-containing 43C |
| chr4_-_43045685 | 0.15 |
ENSMUST00000107956.8
ENSMUST00000107957.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr9_+_107458495 | 0.15 |
ENSMUST00000040059.9
|
Hyal3
|
hyaluronoglucosaminidase 3 |
| chr11_+_66802807 | 0.15 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr17_+_27236961 | 0.15 |
ENSMUST00000142141.3
ENSMUST00000122106.9 |
Ggnbp1
|
gametogenetin binding protein 1 |
| chr8_+_36054919 | 0.15 |
ENSMUST00000037666.6
|
Mfhas1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr16_+_51851588 | 0.15 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr17_-_84773544 | 0.14 |
ENSMUST00000047524.10
|
Thada
|
thyroid adenoma associated |
| chr11_+_115272732 | 0.14 |
ENSMUST00000053288.6
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr15_-_77653979 | 0.13 |
ENSMUST00000229259.2
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr1_+_132243849 | 0.13 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr17_+_48571298 | 0.13 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr19_+_23881821 | 0.13 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr3_+_36917251 | 0.13 |
ENSMUST00000057272.15
|
4932438A13Rik
|
RIKEN cDNA 4932438A13 gene |
| chr15_+_8198506 | 0.13 |
ENSMUST00000110617.2
|
Cplane1
|
ciliogenesis and planar polarity effector 1 |
| chr7_-_80051455 | 0.13 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr18_+_36098090 | 0.13 |
ENSMUST00000176873.8
ENSMUST00000177432.8 ENSMUST00000175734.2 |
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr7_-_12552764 | 0.13 |
ENSMUST00000108546.2
ENSMUST00000072222.8 |
Zfp329
|
zinc finger protein 329 |
| chr7_+_30252687 | 0.13 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr6_+_17307639 | 0.13 |
ENSMUST00000115453.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr4_-_107889267 | 0.13 |
ENSMUST00000106709.9
|
Podn
|
podocan |
| chr16_+_31247562 | 0.12 |
ENSMUST00000115227.10
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr2_-_39080701 | 0.12 |
ENSMUST00000142872.2
ENSMUST00000038874.12 |
Scai
|
suppressor of cancer cell invasion |
| chr9_-_62888156 | 0.12 |
ENSMUST00000098651.6
ENSMUST00000214830.2 |
Pias1
|
protein inhibitor of activated STAT 1 |
| chr9_+_86625694 | 0.12 |
ENSMUST00000179574.2
ENSMUST00000036426.13 |
Prss35
|
protease, serine 35 |
| chr7_-_28078671 | 0.12 |
ENSMUST00000209061.2
|
Zfp36
|
zinc finger protein 36 |
| chr11_-_70350783 | 0.12 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr19_-_29782907 | 0.12 |
ENSMUST00000175726.8
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr13_+_54738014 | 0.11 |
ENSMUST00000026986.7
|
Higd2a
|
HIG1 domain family, member 2A |
| chr6_-_146403638 | 0.11 |
ENSMUST00000079573.13
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr4_-_107889136 | 0.11 |
ENSMUST00000106708.8
|
Podn
|
podocan |
| chr3_+_138058139 | 0.11 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr15_-_99355623 | 0.11 |
ENSMUST00000023747.14
|
Nckap5l
|
NCK-associated protein 5-like |
| chr11_+_46462363 | 0.11 |
ENSMUST00000050937.7
|
BC053393
|
cDNA sequence BC053393 |
| chr12_+_84820024 | 0.11 |
ENSMUST00000021667.7
ENSMUST00000222449.2 ENSMUST00000222982.2 |
Isca2
|
iron-sulfur cluster assembly 2 |
| chr11_-_6556053 | 0.11 |
ENSMUST00000045713.4
|
Nacad
|
NAC alpha domain containing |
| chr1_-_93029532 | 0.11 |
ENSMUST00000171796.8
|
Kif1a
|
kinesin family member 1A |
| chr3_-_126792056 | 0.10 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr7_-_45364613 | 0.10 |
ENSMUST00000072836.7
|
Sphk2
|
sphingosine kinase 2 |
| chr7_-_44888532 | 0.10 |
ENSMUST00000033063.15
|
Cd37
|
CD37 antigen |
| chr6_-_125357756 | 0.10 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr4_+_84802513 | 0.10 |
ENSMUST00000047023.13
|
Cntln
|
centlein, centrosomal protein |
| chr1_-_125839897 | 0.10 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr14_-_52616625 | 0.09 |
ENSMUST00000214980.2
|
Olfr1512
|
olfactory receptor 1512 |
| chr13_-_56444118 | 0.09 |
ENSMUST00000224801.2
|
Cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr11_+_95603494 | 0.09 |
ENSMUST00000107717.8
|
Zfp652
|
zinc finger protein 652 |
| chr8_+_87350672 | 0.09 |
ENSMUST00000034141.18
ENSMUST00000122188.10 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr10_+_98750978 | 0.09 |
ENSMUST00000020107.8
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr4_-_117740624 | 0.09 |
ENSMUST00000030266.12
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr1_+_91294133 | 0.09 |
ENSMUST00000086861.12
|
Erfe
|
erythroferrone |
| chrX_+_99773523 | 0.09 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr1_-_133849131 | 0.09 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr11_-_22236795 | 0.09 |
ENSMUST00000180360.8
ENSMUST00000109563.9 |
Ehbp1
|
EH domain binding protein 1 |
| chr1_-_131172903 | 0.09 |
ENSMUST00000027688.15
ENSMUST00000112442.2 |
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr3_-_20329823 | 0.09 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr7_-_45044606 | 0.09 |
ENSMUST00000209858.2
|
Snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr8_-_106660470 | 0.09 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chrX_+_99773784 | 0.09 |
ENSMUST00000113744.2
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chrX_+_41239872 | 0.08 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2 |
| chr11_-_118233326 | 0.08 |
ENSMUST00000103024.4
|
Cep295nl
|
CEP295 N-terminal like |
| chr2_-_10053381 | 0.08 |
ENSMUST00000026888.11
|
Taf3
|
TATA-box binding protein associated factor 3 |
| chr8_+_84874654 | 0.08 |
ENSMUST00000143833.8
ENSMUST00000118856.8 |
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr10_+_86541675 | 0.08 |
ENSMUST00000075632.14
ENSMUST00000217747.2 ENSMUST00000061458.9 |
Ttc41
|
tetratricopeptide repeat domain 41 |
| chr15_-_83135920 | 0.08 |
ENSMUST00000229687.2
ENSMUST00000164614.3 ENSMUST00000049530.14 |
A4galt
|
alpha 1,4-galactosyltransferase |
| chr8_-_106363982 | 0.08 |
ENSMUST00000182863.2
ENSMUST00000194091.6 |
Gm5914
Agrp
|
predicted gene 5914 agouti related neuropeptide |
| chr7_-_28001624 | 0.08 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr12_+_3476857 | 0.08 |
ENSMUST00000111215.10
ENSMUST00000092003.12 ENSMUST00000144247.9 ENSMUST00000153102.9 |
Asxl2
|
ASXL transcriptional regulator 2 |
| chr3_-_95214443 | 0.08 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr7_+_44398008 | 0.08 |
ENSMUST00000171821.8
|
Vrk3
|
vaccinia related kinase 3 |
| chr17_-_23990512 | 0.07 |
ENSMUST00000226460.2
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr14_-_70855980 | 0.07 |
ENSMUST00000228001.2
|
Dmtn
|
dematin actin binding protein |
| chr2_+_24076481 | 0.07 |
ENSMUST00000057567.3
|
Il1f9
|
interleukin 1 family, member 9 |
| chr11_-_116545071 | 0.07 |
ENSMUST00000021166.6
|
Cygb
|
cytoglobin |
| chr5_-_124564014 | 0.07 |
ENSMUST00000196329.5
ENSMUST00000196644.5 |
Sbno1
|
strawberry notch 1 |
| chr1_+_171157137 | 0.07 |
ENSMUST00000142063.8
ENSMUST00000129116.8 |
Dedd
|
death effector domain-containing |
| chr9_-_45817666 | 0.07 |
ENSMUST00000161187.8
|
Rnf214
|
ring finger protein 214 |
| chr2_-_181333597 | 0.07 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr10_-_127587576 | 0.07 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr17_-_47015928 | 0.07 |
ENSMUST00000002839.9
ENSMUST00000233988.2 |
Ppp2r5d
|
protein phosphatase 2, regulatory subunit B', delta |
| chr4_+_154954042 | 0.07 |
ENSMUST00000079269.14
ENSMUST00000163732.8 ENSMUST00000080559.13 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr17_-_32255287 | 0.06 |
ENSMUST00000238192.2
|
Hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr10_+_84412490 | 0.06 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr7_+_4240697 | 0.06 |
ENSMUST00000117550.2
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr17_+_47815998 | 0.06 |
ENSMUST00000183210.2
|
Ccnd3
|
cyclin D3 |
| chr3_-_89309944 | 0.06 |
ENSMUST00000057431.6
|
Lenep
|
lens epithelial protein |
| chr2_+_121189091 | 0.06 |
ENSMUST00000000317.13
ENSMUST00000129130.3 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr6_+_17307272 | 0.06 |
ENSMUST00000115454.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr3_+_90173813 | 0.06 |
ENSMUST00000098914.10
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr19_-_23251179 | 0.06 |
ENSMUST00000087556.7
ENSMUST00000223934.2 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr5_+_92750898 | 0.06 |
ENSMUST00000200941.2
ENSMUST00000050952.4 |
Stbd1
|
starch binding domain 1 |
| chr6_-_119307685 | 0.06 |
ENSMUST00000112756.8
ENSMUST00000068351.14 |
Lrtm2
|
leucine-rich repeats and transmembrane domains 2 |
| chr19_+_6391148 | 0.06 |
ENSMUST00000025897.13
ENSMUST00000130382.8 |
Map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr14_-_70414236 | 0.06 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr19_-_4861536 | 0.06 |
ENSMUST00000179909.8
ENSMUST00000172000.3 ENSMUST00000180008.2 ENSMUST00000113793.4 ENSMUST00000006625.8 |
Gm21992
Rbm14
|
predicted gene 21992 RNA binding motif protein 14 |
| chr2_+_121188195 | 0.06 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr15_-_78280099 | 0.06 |
ENSMUST00000229878.2
ENSMUST00000165170.8 ENSMUST00000074380.14 |
Tex33
|
testis expressed 33 |
| chr18_-_55123153 | 0.06 |
ENSMUST00000064763.7
|
Zfp608
|
zinc finger protein 608 |
| chr3_-_127019496 | 0.05 |
ENSMUST00000182064.9
ENSMUST00000182452.8 |
Ank2
|
ankyrin 2, brain |
| chrX_+_40490005 | 0.05 |
ENSMUST00000115103.9
ENSMUST00000076349.12 |
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr15_-_78687216 | 0.05 |
ENSMUST00000164826.8
|
Card10
|
caspase recruitment domain family, member 10 |
| chr10_+_43455919 | 0.05 |
ENSMUST00000214476.2
|
Cd24a
|
CD24a antigen |
| chr7_+_89779564 | 0.05 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chrX_-_94240056 | 0.05 |
ENSMUST00000200628.2
ENSMUST00000197364.5 ENSMUST00000181987.8 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr6_-_90405377 | 0.05 |
ENSMUST00000135757.3
ENSMUST00000165673.5 ENSMUST00000062750.12 ENSMUST00000127508.4 |
Cfap100
|
cilia and flagella associated protein 100 |
| chr18_-_60881679 | 0.05 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr19_+_38919477 | 0.05 |
ENSMUST00000145051.2
|
Hells
|
helicase, lymphoid specific |
| chr7_-_74204474 | 0.05 |
ENSMUST00000107453.8
|
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr3_+_75464837 | 0.05 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr1_+_39407183 | 0.05 |
ENSMUST00000195123.6
|
Rpl31
|
ribosomal protein L31 |
| chr8_+_84874881 | 0.05 |
ENSMUST00000093375.5
|
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr3_-_95214102 | 0.05 |
ENSMUST00000107183.8
ENSMUST00000164406.8 ENSMUST00000123365.2 |
Anxa9
|
annexin A9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.7 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.9 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.5 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 0.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.2 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.1 | 0.2 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.3 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.0 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:1903921 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.2 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 1.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.3 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.3 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.5 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0061723 | glycophagy(GO:0061723) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:2000156 | regulation of retrograde vesicle-mediated transport, Golgi to ER(GO:2000156) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.2 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.0 | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.2 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |