Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
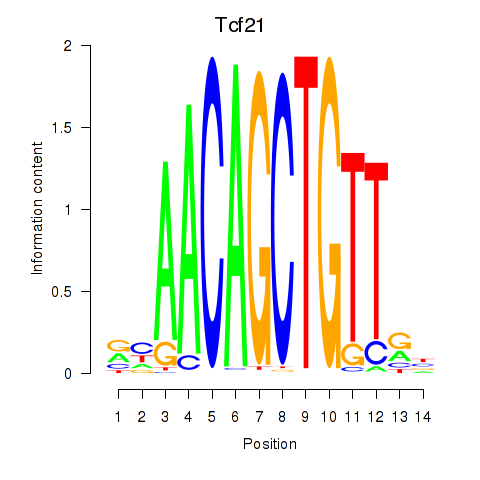
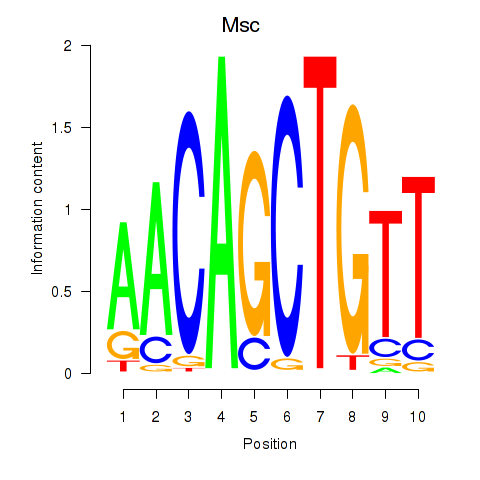
Results for Tcf21_Msc
Z-value: 0.69
Transcription factors associated with Tcf21_Msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf21
|
ENSMUSG00000045680.9 | transcription factor 21 |
|
Msc
|
ENSMUSG00000025930.7 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf21 | mm39_v1_chr10_-_22696025_22696073 | -0.95 | 1.2e-02 | Click! |
| Msc | mm39_v1_chr1_-_14826208_14826222 | -0.40 | 5.1e-01 | Click! |
Activity profile of Tcf21_Msc motif
Sorted Z-values of Tcf21_Msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_122441719 | 0.53 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr11_+_87651359 | 0.50 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr4_-_87724533 | 0.40 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr11_+_53661251 | 0.39 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr13_-_100922910 | 0.38 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr18_+_60509101 | 0.38 |
ENSMUST00000032473.7
ENSMUST00000066912.13 |
Iigp1
|
interferon inducible GTPase 1 |
| chr12_+_24758240 | 0.37 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr13_-_97897139 | 0.36 |
ENSMUST00000074072.5
|
Rps18-ps6
|
ribosomal protein S18, pseudogene 6 |
| chr4_+_152284261 | 0.35 |
ENSMUST00000105652.3
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr10_-_7539009 | 0.33 |
ENSMUST00000163085.8
ENSMUST00000159917.8 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr4_-_87724512 | 0.33 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr2_+_125708027 | 0.33 |
ENSMUST00000134337.8
ENSMUST00000094604.9 |
Galk2
|
galactokinase 2 |
| chr17_-_34218301 | 0.32 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr5_-_123663440 | 0.30 |
ENSMUST00000197682.6
|
Gm49027
|
predicted gene, 49027 |
| chr5_+_123214332 | 0.28 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr10_+_82534841 | 0.28 |
ENSMUST00000020478.14
|
Hcfc2
|
host cell factor C2 |
| chr12_-_72711509 | 0.28 |
ENSMUST00000221750.2
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr5_-_73805063 | 0.28 |
ENSMUST00000081170.9
|
Sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr10_-_7539333 | 0.26 |
ENSMUST00000162606.8
|
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr11_+_117877118 | 0.26 |
ENSMUST00000132676.8
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr13_+_24511387 | 0.25 |
ENSMUST00000224953.2
ENSMUST00000050859.13 ENSMUST00000167746.8 ENSMUST00000224819.2 |
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr8_+_120121612 | 0.25 |
ENSMUST00000098367.5
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr15_+_59186876 | 0.24 |
ENSMUST00000022977.14
ENSMUST00000100640.5 |
Sqle
|
squalene epoxidase |
| chr12_-_72711533 | 0.24 |
ENSMUST00000021512.11
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr11_-_3321307 | 0.24 |
ENSMUST00000101640.10
ENSMUST00000101642.10 |
Limk2
|
LIM motif-containing protein kinase 2 |
| chr7_+_142558783 | 0.23 |
ENSMUST00000009396.13
|
Tspan32
|
tetraspanin 32 |
| chr2_-_174280811 | 0.23 |
ENSMUST00000016400.9
|
Ctsz
|
cathepsin Z |
| chr9_-_114610879 | 0.23 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr7_-_126303887 | 0.23 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr14_-_31299275 | 0.23 |
ENSMUST00000112027.9
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr5_-_138169509 | 0.22 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr7_+_3697511 | 0.22 |
ENSMUST00000108627.4
|
Tsen34
|
tRNA splicing endonuclease subunit 34 |
| chr3_-_105708632 | 0.22 |
ENSMUST00000090678.11
|
Rap1a
|
RAS-related protein 1a |
| chr2_-_25114660 | 0.22 |
ENSMUST00000043584.5
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr3_-_105708601 | 0.22 |
ENSMUST00000197094.5
ENSMUST00000198004.2 |
Rap1a
|
RAS-related protein 1a |
| chr16_+_44632096 | 0.22 |
ENSMUST00000176819.8
ENSMUST00000176321.8 |
Cd200r4
|
CD200 receptor 4 |
| chr4_-_132990362 | 0.22 |
ENSMUST00000105908.10
ENSMUST00000030674.8 |
Sytl1
|
synaptotagmin-like 1 |
| chr3_+_121243395 | 0.22 |
ENSMUST00000198393.2
|
Cnn3
|
calponin 3, acidic |
| chr3_+_14928561 | 0.21 |
ENSMUST00000029076.6
|
Car3
|
carbonic anhydrase 3 |
| chr8_-_108151661 | 0.21 |
ENSMUST00000003946.9
|
Nob1
|
NIN1/RPN12 binding protein 1 homolog |
| chr16_-_4377640 | 0.21 |
ENSMUST00000005862.9
|
Tfap4
|
transcription factor AP4 |
| chr12_+_24758724 | 0.21 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr4_+_133970973 | 0.21 |
ENSMUST00000135228.8
ENSMUST00000144222.8 ENSMUST00000143448.8 ENSMUST00000125921.8 ENSMUST00000122952.8 ENSMUST00000131447.2 |
E130218I03Rik
|
RIKEN cDNA E130218I03 gene |
| chr11_-_74615496 | 0.21 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr7_-_37718916 | 0.21 |
ENSMUST00000085513.6
ENSMUST00000206327.2 |
Uri1
|
URI1, prefoldin-like chaperone |
| chr13_-_47168286 | 0.21 |
ENSMUST00000052747.4
|
Nhlrc1
|
NHL repeat containing 1 |
| chr3_+_95496270 | 0.21 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr7_-_126303689 | 0.20 |
ENSMUST00000135087.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr6_+_145091196 | 0.20 |
ENSMUST00000156849.8
ENSMUST00000132948.2 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr13_-_33035150 | 0.20 |
ENSMUST00000091668.13
ENSMUST00000076352.8 |
Serpinb1a
|
serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| chr7_+_142558837 | 0.20 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chr16_+_49620883 | 0.20 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr5_+_64317550 | 0.20 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr3_+_19241484 | 0.20 |
ENSMUST00000130806.8
ENSMUST00000117529.8 ENSMUST00000119865.8 |
Mtfr1
|
mitochondrial fission regulator 1 |
| chr5_+_35146727 | 0.19 |
ENSMUST00000114284.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr7_-_141579689 | 0.19 |
ENSMUST00000209732.2
|
Mob2
|
MOB kinase activator 2 |
| chr11_-_48836975 | 0.19 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chrX_+_108138965 | 0.18 |
ENSMUST00000033598.9
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr3_-_94949854 | 0.18 |
ENSMUST00000117355.2
ENSMUST00000071664.12 ENSMUST00000107237.8 |
Psmd4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| chr17_-_24926589 | 0.18 |
ENSMUST00000126319.8
|
Tbl3
|
transducin (beta)-like 3 |
| chr19_+_11493825 | 0.18 |
ENSMUST00000163078.8
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr3_-_129625023 | 0.18 |
ENSMUST00000029643.15
|
Gar1
|
GAR1 ribonucleoprotein |
| chrX_+_74557905 | 0.18 |
ENSMUST00000114070.10
ENSMUST00000033540.6 |
Vbp1
|
von Hippel-Lindau binding protein 1 |
| chr14_+_55716197 | 0.18 |
ENSMUST00000022821.8
|
Dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr11_-_99045894 | 0.18 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr2_+_130116357 | 0.17 |
ENSMUST00000136621.9
ENSMUST00000141872.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr7_-_127494750 | 0.17 |
ENSMUST00000033074.8
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr2_-_35322581 | 0.17 |
ENSMUST00000079424.11
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr6_+_17749169 | 0.17 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr8_-_71834543 | 0.17 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr11_+_5470968 | 0.17 |
ENSMUST00000239150.2
|
Xbp1
|
X-box binding protein 1 |
| chr14_+_55716227 | 0.16 |
ENSMUST00000227488.2
|
Dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr13_+_21364330 | 0.16 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr3_+_106393348 | 0.16 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr19_-_23425757 | 0.16 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chr17_-_36432041 | 0.16 |
ENSMUST00000166442.3
|
H2-T10
|
histocompatibility 2, T region locus 10 |
| chr3_+_40755211 | 0.15 |
ENSMUST00000204473.2
|
Plk4
|
polo like kinase 4 |
| chr2_-_28730286 | 0.15 |
ENSMUST00000037117.6
ENSMUST00000171404.8 |
Gtf3c4
|
general transcription factor IIIC, polypeptide 4 |
| chr7_+_140343652 | 0.15 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr10_+_82821304 | 0.15 |
ENSMUST00000040110.8
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr11_-_55310724 | 0.15 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr11_+_101207743 | 0.15 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr13_-_4329421 | 0.15 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr5_-_138170077 | 0.15 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_121590524 | 0.15 |
ENSMUST00000052590.8
ENSMUST00000130451.2 |
Erp29
|
endoplasmic reticulum protein 29 |
| chr15_+_102379503 | 0.15 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr3_-_53771185 | 0.15 |
ENSMUST00000122330.2
ENSMUST00000146598.8 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr19_-_15901919 | 0.14 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr19_+_8779903 | 0.14 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chrX_-_70536198 | 0.14 |
ENSMUST00000080035.11
|
Cd99l2
|
CD99 antigen-like 2 |
| chr15_+_84208980 | 0.14 |
ENSMUST00000145809.2
|
Parvg
|
parvin, gamma |
| chr8_+_106428256 | 0.14 |
ENSMUST00000093195.7
ENSMUST00000211888.2 ENSMUST00000212430.2 |
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr5_-_140375010 | 0.14 |
ENSMUST00000031539.12
|
Snx8
|
sorting nexin 8 |
| chr13_+_52000704 | 0.14 |
ENSMUST00000021903.3
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr7_-_51655426 | 0.14 |
ENSMUST00000209193.2
|
Svip
|
small VCP/p97-interacting protein |
| chr15_+_34238174 | 0.13 |
ENSMUST00000022867.5
ENSMUST00000226627.2 |
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chrX_+_35592006 | 0.13 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr5_-_52628825 | 0.13 |
ENSMUST00000198008.5
ENSMUST00000059428.7 |
Ccdc149
|
coiled-coil domain containing 149 |
| chr2_+_140012560 | 0.13 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr11_-_76289888 | 0.13 |
ENSMUST00000021204.4
|
Nxn
|
nucleoredoxin |
| chr17_+_28059129 | 0.13 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr9_-_103940247 | 0.13 |
ENSMUST00000035166.12
|
Uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr10_+_127871444 | 0.13 |
ENSMUST00000073868.9
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr9_+_103940879 | 0.13 |
ENSMUST00000047799.13
ENSMUST00000189998.3 |
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr5_-_143951671 | 0.12 |
ENSMUST00000031621.11
|
Ccz1
|
CCZ1 vacuolar protein trafficking and biogenesis associated |
| chr14_-_31552335 | 0.12 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr10_+_75768964 | 0.12 |
ENSMUST00000219839.2
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr4_-_43031370 | 0.12 |
ENSMUST00000138030.2
ENSMUST00000136326.8 |
Stoml2
|
stomatin (Epb7.2)-like 2 |
| chr14_+_55716302 | 0.12 |
ENSMUST00000226168.2
|
Dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr7_-_4517367 | 0.12 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr15_+_41694317 | 0.12 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr3_+_84832783 | 0.12 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr16_-_10265204 | 0.12 |
ENSMUST00000051118.7
|
Tvp23a
|
trans-golgi network vesicle protein 23A |
| chr9_+_65370077 | 0.12 |
ENSMUST00000215170.2
|
Spg21
|
SPG21, maspardin |
| chr8_-_25592001 | 0.12 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr2_+_155223728 | 0.12 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr11_-_84807164 | 0.12 |
ENSMUST00000103195.5
|
Znhit3
|
zinc finger, HIT type 3 |
| chr5_+_91079068 | 0.12 |
ENSMUST00000202781.2
ENSMUST00000071652.6 |
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr5_+_147456497 | 0.11 |
ENSMUST00000175807.8
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr8_-_111629074 | 0.11 |
ENSMUST00000041382.7
|
Fcsk
|
fucose kinase |
| chr11_-_31320065 | 0.11 |
ENSMUST00000020546.3
|
Stc2
|
stanniocalcin 2 |
| chr16_-_45975440 | 0.11 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr6_-_76474767 | 0.11 |
ENSMUST00000097218.7
|
Gm9008
|
predicted pseudogene 9008 |
| chr11_+_49141339 | 0.11 |
ENSMUST00000101293.11
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr1_-_182929025 | 0.11 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr1_-_53391778 | 0.11 |
ENSMUST00000236737.2
ENSMUST00000027264.10 ENSMUST00000123519.9 |
Gm50478
Asnsd1
|
predicted gene, 50478 asparagine synthetase domain containing 1 |
| chr7_-_79882501 | 0.11 |
ENSMUST00000065163.15
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr9_+_103940575 | 0.11 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr7_-_110673269 | 0.11 |
ENSMUST00000163014.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr4_-_133694607 | 0.11 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_-_52271455 | 0.11 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr1_-_16689527 | 0.11 |
ENSMUST00000182554.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr4_+_144619696 | 0.11 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr18_+_48178397 | 0.11 |
ENSMUST00000076155.6
|
Eno1b
|
enolase 1B, retrotransposed |
| chr11_+_32155483 | 0.11 |
ENSMUST00000121182.2
|
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr2_-_73044667 | 0.11 |
ENSMUST00000100015.6
|
Ola1
|
Obg-like ATPase 1 |
| chr11_+_32155415 | 0.11 |
ENSMUST00000039601.10
ENSMUST00000149043.3 |
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr9_+_63509925 | 0.11 |
ENSMUST00000041551.9
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr8_-_85573489 | 0.11 |
ENSMUST00000003912.7
|
Calr
|
calreticulin |
| chr15_+_76460550 | 0.10 |
ENSMUST00000162503.8
|
Adck5
|
aarF domain containing kinase 5 |
| chr2_+_29968596 | 0.10 |
ENSMUST00000045246.8
|
Pkn3
|
protein kinase N3 |
| chr3_+_95496239 | 0.10 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr1_+_162398256 | 0.10 |
ENSMUST00000194810.6
ENSMUST00000050010.11 ENSMUST00000150040.8 |
Vamp4
|
vesicle-associated membrane protein 4 |
| chr13_-_8921732 | 0.10 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr17_-_25104874 | 0.10 |
ENSMUST00000234597.2
|
Nubp2
|
nucleotide binding protein 2 |
| chr3_-_88857213 | 0.10 |
ENSMUST00000172942.8
ENSMUST00000107491.11 |
Dap3
|
death associated protein 3 |
| chr12_-_40087393 | 0.10 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr15_-_98816012 | 0.10 |
ENSMUST00000023736.10
|
Lmbr1l
|
limb region 1 like |
| chr14_-_56809287 | 0.10 |
ENSMUST00000065302.8
|
Cenpj
|
centromere protein J |
| chr9_+_121539395 | 0.10 |
ENSMUST00000035113.11
ENSMUST00000215966.2 ENSMUST00000215833.2 ENSMUST00000215104.2 |
Ss18l2
|
SS18, nBAF chromatin remodeling complex subunit like 2 |
| chr9_-_24414423 | 0.10 |
ENSMUST00000142064.8
ENSMUST00000170356.2 |
Dpy19l1
|
dpy-19-like 1 (C. elegans) |
| chr2_-_7400690 | 0.10 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr11_-_69576363 | 0.10 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr6_-_29212295 | 0.10 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chrX_-_100310959 | 0.10 |
ENSMUST00000135038.2
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr1_-_36574801 | 0.10 |
ENSMUST00000054665.10
ENSMUST00000194853.3 |
Ankrd23
|
ankyrin repeat domain 23 |
| chr8_+_85583611 | 0.10 |
ENSMUST00000003906.13
ENSMUST00000109754.2 |
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr1_+_128069716 | 0.10 |
ENSMUST00000187557.2
|
R3hdm1
|
R3H domain containing 1 |
| chr5_-_122639840 | 0.10 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr6_-_29212239 | 0.10 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr5_-_73349191 | 0.09 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr4_+_97665843 | 0.09 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr17_-_75858835 | 0.09 |
ENSMUST00000234785.2
ENSMUST00000112507.4 |
Fam98a
|
family with sequence similarity 98, member A |
| chr7_-_51655542 | 0.09 |
ENSMUST00000098414.5
|
Svip
|
small VCP/p97-interacting protein |
| chr7_-_30855295 | 0.09 |
ENSMUST00000186723.3
|
Gramd1a
|
GRAM domain containing 1A |
| chr5_+_138170259 | 0.09 |
ENSMUST00000019662.11
ENSMUST00000151318.8 |
Ap4m1
|
adaptor-related protein complex AP-4, mu 1 |
| chr14_-_55114989 | 0.09 |
ENSMUST00000168622.2
ENSMUST00000177403.2 |
Ppp1r3e
|
protein phosphatase 1, regulatory subunit 3E |
| chr2_+_32647246 | 0.09 |
ENSMUST00000009707.14
ENSMUST00000177382.2 ENSMUST00000140999.2 |
Tor2a
|
torsin family 2, member A |
| chr4_-_135699205 | 0.09 |
ENSMUST00000105852.8
|
Lypla2
|
lysophospholipase 2 |
| chr19_-_55087849 | 0.09 |
ENSMUST00000061856.6
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr8_-_79539838 | 0.09 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_+_55170411 | 0.09 |
ENSMUST00000027122.14
|
Mob4
|
MOB family member 4, phocein |
| chrX_+_73473277 | 0.09 |
ENSMUST00000114127.8
ENSMUST00000064407.10 ENSMUST00000156707.3 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr4_+_46489248 | 0.09 |
ENSMUST00000030018.5
|
Nans
|
N-acetylneuraminic acid synthase (sialic acid synthase) |
| chr11_+_99748741 | 0.09 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr4_-_42665763 | 0.09 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr5_+_36622342 | 0.09 |
ENSMUST00000031099.4
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr11_+_82845382 | 0.09 |
ENSMUST00000108157.2
|
Slfn5
|
schlafen 5 |
| chr19_+_4560500 | 0.09 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr4_-_42168603 | 0.09 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr15_+_102379621 | 0.09 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr6_+_58573656 | 0.09 |
ENSMUST00000031822.13
|
Abcg2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr16_+_23338960 | 0.08 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr4_+_15265798 | 0.08 |
ENSMUST00000062684.9
|
Tmem64
|
transmembrane protein 64 |
| chr7_-_127593003 | 0.08 |
ENSMUST00000033056.5
|
Pycard
|
PYD and CARD domain containing |
| chr9_+_57444801 | 0.08 |
ENSMUST00000093833.6
ENSMUST00000114200.10 |
Fam219b
|
family with sequence similarity 219, member B |
| chrX_+_55500170 | 0.08 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr2_-_32737208 | 0.08 |
ENSMUST00000077458.7
ENSMUST00000208840.2 |
Stxbp1
|
syntaxin binding protein 1 |
| chr5_+_25452342 | 0.08 |
ENSMUST00000114950.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr12_-_112905721 | 0.08 |
ENSMUST00000221497.2
ENSMUST00000002881.5 ENSMUST00000221397.2 |
Nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr15_-_77854711 | 0.08 |
ENSMUST00000230419.2
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr9_-_40257586 | 0.08 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr17_-_66408461 | 0.08 |
ENSMUST00000024909.15
ENSMUST00000147484.2 |
Ndufv2
|
NADH:ubiquinone oxidoreductase core subunit V2 |
| chr15_-_89007290 | 0.08 |
ENSMUST00000109353.9
|
Tubgcp6
|
tubulin, gamma complex associated protein 6 |
| chr8_+_106412905 | 0.08 |
ENSMUST00000213019.2
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr9_-_22218934 | 0.08 |
ENSMUST00000086278.7
ENSMUST00000215202.2 |
Zfp810
|
zinc finger protein 810 |
| chr3_+_89344006 | 0.08 |
ENSMUST00000038942.10
ENSMUST00000130858.8 ENSMUST00000146630.8 ENSMUST00000145753.2 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
| chr15_+_9071761 | 0.08 |
ENSMUST00000189437.8
ENSMUST00000190874.8 |
Nadk2
|
NAD kinase 2, mitochondrial |
| chr2_+_32126591 | 0.08 |
ENSMUST00000036473.16
|
Pomt1
|
protein-O-mannosyltransferase 1 |
| chr11_-_20781009 | 0.08 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf21_Msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.6 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.1 | 0.4 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.2 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.1 | 0.4 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 0.3 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 0.3 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.2 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.4 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.4 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.2 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.6 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.4 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.2 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.1 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.0 | 0.1 | GO:0002582 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 | 0.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0002191 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.0 | 0.1 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.0 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.2 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.4 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.4 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.1 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.4 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.0 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.5 | GO:0018455 | alcohol dehydrogenase [NAD(P)+] activity(GO:0018455) |
| 0.1 | 0.6 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.1 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.2 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.0 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |