Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Tcf7l1
Z-value: 0.35
Transcription factors associated with Tcf7l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf7l1
|
ENSMUSG00000055799.14 | transcription factor 7 like 1 (T cell specific, HMG box) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7l1 | mm39_v1_chr6_-_72766224_72766245 | -0.70 | 1.9e-01 | Click! |
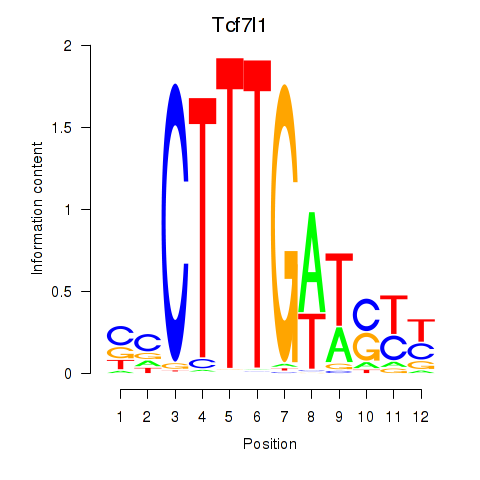
Activity profile of Tcf7l1 motif
Sorted Z-values of Tcf7l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_65288418 | 0.28 |
ENSMUST00000101191.10
ENSMUST00000204348.3 |
Klhl5
|
kelch-like 5 |
| chr6_-_52217821 | 0.26 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr3_-_50398027 | 0.25 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr7_-_117728790 | 0.25 |
ENSMUST00000206491.2
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr7_-_144493560 | 0.24 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr14_-_98406977 | 0.21 |
ENSMUST00000071533.13
ENSMUST00000069334.8 |
Dach1
|
dachshund family transcription factor 1 |
| chr17_-_90217868 | 0.18 |
ENSMUST00000086423.6
|
Gm10184
|
predicted pseudogene 10184 |
| chr5_+_31350607 | 0.15 |
ENSMUST00000201535.4
|
Snx17
|
sorting nexin 17 |
| chr7_+_27290969 | 0.15 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr3_-_122413361 | 0.14 |
ENSMUST00000239148.2
ENSMUST00000162409.8 ENSMUST00000162947.3 |
Fnbp1l
|
formin binding protein 1-like |
| chr6_+_48578935 | 0.13 |
ENSMUST00000204095.3
|
Zfp775
|
zinc finger protein 775 |
| chr6_-_47790272 | 0.13 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr13_-_119545479 | 0.13 |
ENSMUST00000223268.2
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr15_-_98851566 | 0.13 |
ENSMUST00000097014.7
|
Tuba1a
|
tubulin, alpha 1A |
| chr5_-_148329615 | 0.12 |
ENSMUST00000138257.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr10_+_96453408 | 0.12 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr11_-_61652866 | 0.12 |
ENSMUST00000004955.14
ENSMUST00000168115.8 |
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr6_-_52211882 | 0.12 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr16_-_16950241 | 0.12 |
ENSMUST00000023453.10
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr1_+_131080912 | 0.12 |
ENSMUST00000112446.9
ENSMUST00000068805.14 ENSMUST00000068791.11 |
Eif2d
|
eukaryotic translation initiation factor 2D |
| chr13_-_30168374 | 0.12 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
| chr13_-_119545520 | 0.12 |
ENSMUST00000069902.13
ENSMUST00000099149.10 ENSMUST00000109204.8 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr19_-_46327024 | 0.12 |
ENSMUST00000236046.2
ENSMUST00000236980.2 ENSMUST00000235485.2 ENSMUST00000236061.2 ENSMUST00000236236.2 ENSMUST00000236768.2 ENSMUST00000236651.2 |
Cuedc2
|
CUE domain containing 2 |
| chr7_+_27291126 | 0.12 |
ENSMUST00000167435.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr11_-_48836975 | 0.11 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr7_-_117728815 | 0.11 |
ENSMUST00000032888.10
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr19_-_46327071 | 0.11 |
ENSMUST00000235977.2
ENSMUST00000167861.8 ENSMUST00000051234.9 ENSMUST00000236066.2 |
Cuedc2
|
CUE domain containing 2 |
| chr17_-_71158184 | 0.11 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr10_+_79986280 | 0.11 |
ENSMUST00000153477.8
|
Midn
|
midnolin |
| chr13_-_6686686 | 0.11 |
ENSMUST00000136585.2
|
Pfkp
|
phosphofructokinase, platelet |
| chr2_-_69542805 | 0.10 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chrX_+_168662592 | 0.10 |
ENSMUST00000112105.8
ENSMUST00000078947.12 |
Mid1
|
midline 1 |
| chr5_-_31350352 | 0.10 |
ENSMUST00000202758.4
ENSMUST00000114603.8 |
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr3_-_89905547 | 0.10 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr7_+_65759198 | 0.10 |
ENSMUST00000036372.8
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr17_-_71158052 | 0.10 |
ENSMUST00000186358.6
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr12_-_84455764 | 0.10 |
ENSMUST00000120942.8
ENSMUST00000110272.9 |
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr12_-_21467437 | 0.09 |
ENSMUST00000103002.8
ENSMUST00000155480.9 ENSMUST00000135088.9 |
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| chr5_+_31350566 | 0.09 |
ENSMUST00000031029.15
ENSMUST00000201679.4 |
Snx17
|
sorting nexin 17 |
| chr7_-_143102792 | 0.09 |
ENSMUST00000072727.7
ENSMUST00000207948.2 ENSMUST00000208190.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr11_+_3280771 | 0.09 |
ENSMUST00000136536.8
ENSMUST00000093399.11 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_+_32762656 | 0.09 |
ENSMUST00000029214.14
|
Actl6a
|
actin-like 6A |
| chr10_+_53472853 | 0.09 |
ENSMUST00000219271.2
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr6_-_47571901 | 0.08 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr3_-_143910926 | 0.08 |
ENSMUST00000120539.8
ENSMUST00000196264.5 |
Lmo4
|
LIM domain only 4 |
| chr8_-_69636825 | 0.08 |
ENSMUST00000185176.8
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr3_+_32763313 | 0.08 |
ENSMUST00000126144.3
|
Actl6a
|
actin-like 6A |
| chr3_+_135531548 | 0.08 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr9_-_57552392 | 0.08 |
ENSMUST00000216934.2
|
Csk
|
c-src tyrosine kinase |
| chr2_+_18677195 | 0.08 |
ENSMUST00000171845.8
ENSMUST00000061158.5 |
Commd3
|
COMM domain containing 3 |
| chr12_-_31401432 | 0.08 |
ENSMUST00000110857.5
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr2_-_35322581 | 0.08 |
ENSMUST00000079424.11
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr2_+_163500290 | 0.07 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr6_-_52195663 | 0.07 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr14_+_71011744 | 0.07 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr2_+_26205525 | 0.07 |
ENSMUST00000066936.9
ENSMUST00000078616.12 |
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr11_+_108811626 | 0.07 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr10_+_53473032 | 0.07 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr1_-_182929025 | 0.07 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr1_+_165616250 | 0.07 |
ENSMUST00000161971.8
ENSMUST00000187313.7 ENSMUST00000178336.8 ENSMUST00000005907.12 ENSMUST00000027849.11 |
Cd247
|
CD247 antigen |
| chr15_-_37007623 | 0.07 |
ENSMUST00000078976.9
|
Zfp706
|
zinc finger protein 706 |
| chr4_+_148215339 | 0.07 |
ENSMUST00000084129.9
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr9_+_65583826 | 0.07 |
ENSMUST00000153700.9
ENSMUST00000046490.14 |
Oaz2
|
ornithine decarboxylase antizyme 2 |
| chr11_+_80367839 | 0.07 |
ENSMUST00000053413.12
ENSMUST00000147694.2 |
Cdk5r1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr3_+_84081411 | 0.07 |
ENSMUST00000193882.2
|
Gm6525
|
predicted pseudogene 6525 |
| chr11_+_69737491 | 0.07 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr12_+_84455813 | 0.06 |
ENSMUST00000081828.13
|
Bbof1
|
basal body orientation factor 1 |
| chr11_+_69737200 | 0.06 |
ENSMUST00000108632.8
|
Plscr3
|
phospholipid scramblase 3 |
| chr4_+_108336164 | 0.06 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr10_-_118179160 | 0.06 |
ENSMUST00000218329.2
|
Gm9046
|
predicted gene 9046 |
| chr7_+_82516491 | 0.06 |
ENSMUST00000082237.7
|
Mex3b
|
mex3 RNA binding family member B |
| chr4_+_11758147 | 0.06 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr14_-_52258158 | 0.06 |
ENSMUST00000228580.2
ENSMUST00000226554.2 ENSMUST00000067549.15 |
Zfp219
|
zinc finger protein 219 |
| chr11_+_108811168 | 0.06 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr12_-_112640626 | 0.06 |
ENSMUST00000001780.10
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chrX_-_149595873 | 0.06 |
ENSMUST00000131241.2
ENSMUST00000147152.3 ENSMUST00000143843.8 |
Maged2
|
MAGE family member D2 |
| chr19_-_47907705 | 0.06 |
ENSMUST00000095998.7
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr3_-_148696155 | 0.06 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr6_+_136305516 | 0.06 |
ENSMUST00000204966.2
ENSMUST00000077886.7 |
Eif4a3l1
|
eukaryotic translation initiation factor 4A3 like 1 |
| chr11_-_49603501 | 0.06 |
ENSMUST00000020624.7
ENSMUST00000145353.8 |
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr11_+_120382666 | 0.06 |
ENSMUST00000026899.4
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
| chr17_-_34250616 | 0.06 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr8_-_4309257 | 0.06 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr13_-_55635851 | 0.06 |
ENSMUST00000109921.9
ENSMUST00000109923.9 ENSMUST00000021950.15 |
Dbn1
|
drebrin 1 |
| chr1_-_58735106 | 0.05 |
ENSMUST00000055313.14
ENSMUST00000188772.7 |
Flacc1
|
flagellum associated containing coiled-coil domains 1 |
| chr1_-_37580084 | 0.05 |
ENSMUST00000151952.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr11_-_6224830 | 0.05 |
ENSMUST00000132147.3
ENSMUST00000004508.13 |
Tmed4
|
transmembrane p24 trafficking protein 4 |
| chr12_-_83643883 | 0.05 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr1_-_45964730 | 0.05 |
ENSMUST00000027137.11
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr18_-_12952925 | 0.05 |
ENSMUST00000119043.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr8_-_86159398 | 0.05 |
ENSMUST00000047749.7
|
4921524J17Rik
|
RIKEN cDNA 4921524J17 gene |
| chr2_+_35472841 | 0.05 |
ENSMUST00000135741.8
|
Dab2ip
|
disabled 2 interacting protein |
| chr13_+_94219934 | 0.05 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr13_-_54737857 | 0.05 |
ENSMUST00000148222.2
ENSMUST00000026987.12 |
Nop16
|
NOP16 nucleolar protein |
| chr9_-_64928927 | 0.05 |
ENSMUST00000036615.7
|
Hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr5_-_33812830 | 0.05 |
ENSMUST00000200849.2
|
Tmem129
|
transmembrane protein 129 |
| chr5_-_31350449 | 0.05 |
ENSMUST00000166769.8
|
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr8_-_61407760 | 0.05 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr11_-_78388560 | 0.05 |
ENSMUST00000061174.7
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr19_+_10019023 | 0.05 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr2_-_164876690 | 0.05 |
ENSMUST00000122070.2
ENSMUST00000121377.8 ENSMUST00000153905.2 ENSMUST00000040381.15 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr10_-_118168313 | 0.05 |
ENSMUST00000179851.3
|
Gm9044
|
predicted gene 9044 |
| chr2_+_160573604 | 0.05 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr7_-_143102524 | 0.05 |
ENSMUST00000208093.2
ENSMUST00000209098.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr4_-_126362372 | 0.05 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr10_-_22607136 | 0.05 |
ENSMUST00000238910.2
ENSMUST00000127698.8 |
ENSMUSG00000118528.2
Tbpl1
|
novel protein TATA box binding protein-like 1 |
| chr8_+_108162985 | 0.05 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr8_-_87472562 | 0.05 |
ENSMUST00000045296.6
|
Siah1a
|
siah E3 ubiquitin protein ligase 1A |
| chr15_-_37008011 | 0.05 |
ENSMUST00000226671.2
|
Zfp706
|
zinc finger protein 706 |
| chr8_+_22996233 | 0.04 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr11_-_101193106 | 0.04 |
ENSMUST00000130916.8
|
Becn1
|
beclin 1, autophagy related |
| chr7_-_83533497 | 0.04 |
ENSMUST00000094216.5
|
Tlnrd1
|
talin rod domain containing 1 |
| chrX_-_56438322 | 0.04 |
ENSMUST00000114730.8
|
Rbmx
|
RNA binding motif protein, X chromosome |
| chr17_+_29712008 | 0.04 |
ENSMUST00000234665.2
|
Pim1
|
proviral integration site 1 |
| chr17_+_28910393 | 0.04 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr11_-_78388284 | 0.04 |
ENSMUST00000108287.10
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr13_+_22129246 | 0.04 |
ENSMUST00000176511.8
ENSMUST00000102978.8 ENSMUST00000152258.9 |
Zfp184
|
zinc finger protein 184 (Kruppel-like) |
| chr16_+_31482949 | 0.04 |
ENSMUST00000023454.12
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr5_+_36050663 | 0.04 |
ENSMUST00000064571.11
|
Afap1
|
actin filament associated protein 1 |
| chr11_-_101193032 | 0.04 |
ENSMUST00000140706.8
ENSMUST00000170502.2 ENSMUST00000172233.8 |
Becn1
|
beclin 1, autophagy related |
| chrX_-_149595711 | 0.04 |
ENSMUST00000112697.10
|
Maged2
|
MAGE family member D2 |
| chr6_+_17065141 | 0.04 |
ENSMUST00000115467.11
ENSMUST00000154266.3 ENSMUST00000076654.9 |
Tes
|
testin LIM domain protein |
| chr17_-_34822649 | 0.04 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr9_+_69896748 | 0.04 |
ENSMUST00000034754.12
ENSMUST00000085393.13 ENSMUST00000117450.8 |
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr4_+_95445731 | 0.04 |
ENSMUST00000079223.11
ENSMUST00000177394.8 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr3_-_89905927 | 0.04 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr1_-_13061333 | 0.04 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr4_-_141391406 | 0.04 |
ENSMUST00000084203.11
|
Plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chrX_-_56438380 | 0.04 |
ENSMUST00000143310.2
ENSMUST00000098470.9 ENSMUST00000114726.8 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr6_-_148846247 | 0.04 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr12_-_83643964 | 0.04 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr13_+_51254852 | 0.03 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chrX_+_158771429 | 0.03 |
ENSMUST00000033665.9
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr11_+_108814007 | 0.03 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chr7_+_24310738 | 0.03 |
ENSMUST00000073325.6
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr8_+_31601837 | 0.03 |
ENSMUST00000046941.8
ENSMUST00000217278.2 |
Rnf122
|
ring finger protein 122 |
| chr6_+_86605146 | 0.03 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr1_+_165616315 | 0.03 |
ENSMUST00000161559.3
|
Cd247
|
CD247 antigen |
| chr18_-_36330367 | 0.03 |
ENSMUST00000115713.9
|
Nrg2
|
neuregulin 2 |
| chr5_+_33978035 | 0.03 |
ENSMUST00000075812.11
ENSMUST00000114397.9 ENSMUST00000155880.8 |
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr6_+_88879233 | 0.03 |
ENSMUST00000055022.15
ENSMUST00000204765.3 ENSMUST00000153874.8 |
Tpra1
|
transmembrane protein, adipocyte asscociated 1 |
| chr9_-_57552844 | 0.03 |
ENSMUST00000216979.2
ENSMUST00000034863.8 |
Csk
|
c-src tyrosine kinase |
| chr2_+_160573178 | 0.03 |
ENSMUST00000103115.8
|
Plcg1
|
phospholipase C, gamma 1 |
| chr2_-_65068960 | 0.03 |
ENSMUST00000112429.9
ENSMUST00000102726.8 ENSMUST00000112430.8 |
Cobll1
|
Cobl-like 1 |
| chr2_+_152873772 | 0.03 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr2_-_114031897 | 0.03 |
ENSMUST00000050668.4
|
Zfp770
|
zinc finger protein 770 |
| chr8_+_79755194 | 0.03 |
ENSMUST00000119254.8
ENSMUST00000238669.2 |
Zfp827
|
zinc finger protein 827 |
| chr10_-_82458760 | 0.03 |
ENSMUST00000176200.8
ENSMUST00000183416.2 |
1190007I07Rik
|
RIKEN cDNA 1190007I07 gene |
| chr6_+_71684846 | 0.03 |
ENSMUST00000212792.2
|
Reep1
|
receptor accessory protein 1 |
| chrX_+_149981074 | 0.02 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr19_+_8816663 | 0.02 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr18_+_61688329 | 0.02 |
ENSMUST00000165123.8
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr9_+_54771064 | 0.02 |
ENSMUST00000034843.9
|
Ireb2
|
iron responsive element binding protein 2 |
| chr2_-_6726701 | 0.02 |
ENSMUST00000114927.9
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr11_-_53959758 | 0.02 |
ENSMUST00000093109.11
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr11_-_3599673 | 0.02 |
ENSMUST00000193809.2
|
Tug1
|
taurine upregulated gene 1 |
| chr12_+_86999366 | 0.02 |
ENSMUST00000191463.2
|
Cipc
|
CLOCK interacting protein, circadian |
| chr17_+_8144822 | 0.02 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr13_-_103042554 | 0.02 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr16_+_93574129 | 0.02 |
ENSMUST00000228261.2
|
Dop1b
|
DOP1 leucine zipper like protein B |
| chr14_+_49409659 | 0.02 |
ENSMUST00000153488.9
|
Naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr12_-_112640378 | 0.02 |
ENSMUST00000130342.2
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr9_+_35334878 | 0.02 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr17_+_37504783 | 0.02 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr3_+_20011405 | 0.02 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr4_+_124923785 | 0.02 |
ENSMUST00000030684.8
|
Gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr1_-_4479445 | 0.02 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr18_-_88912446 | 0.02 |
ENSMUST00000070116.12
ENSMUST00000125362.8 |
Socs6
|
suppressor of cytokine signaling 6 |
| chr14_-_20026761 | 0.02 |
ENSMUST00000161247.2
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr3_+_20011251 | 0.02 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr7_+_73025243 | 0.02 |
ENSMUST00000119206.3
ENSMUST00000094312.12 |
Rgma
|
repulsive guidance molecule family member A |
| chr13_-_99062882 | 0.02 |
ENSMUST00000109401.8
|
Tnpo1
|
transportin 1 |
| chr1_+_36806015 | 0.02 |
ENSMUST00000185871.2
|
Zap70
|
zeta-chain (TCR) associated protein kinase |
| chrX_-_47123719 | 0.02 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr10_+_126836578 | 0.02 |
ENSMUST00000026500.12
ENSMUST00000142698.8 |
Avil
|
advillin |
| chr17_+_34823236 | 0.02 |
ENSMUST00000174041.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr8_+_45960931 | 0.02 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_+_13579092 | 0.02 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr12_-_51876282 | 0.01 |
ENSMUST00000042052.9
ENSMUST00000179265.8 |
Hectd1
|
HECT domain E3 ubiquitin protein ligase 1 |
| chr7_-_110681402 | 0.01 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr3_-_95014090 | 0.01 |
ENSMUST00000005768.8
ENSMUST00000107232.9 ENSMUST00000107236.9 |
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr11_+_108812474 | 0.01 |
ENSMUST00000144511.2
|
Axin2
|
axin 2 |
| chr11_+_69737437 | 0.01 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr9_+_90045219 | 0.01 |
ENSMUST00000147250.8
ENSMUST00000113060.3 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr11_-_53959790 | 0.01 |
ENSMUST00000018755.10
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr3_-_53370621 | 0.01 |
ENSMUST00000056749.14
|
Nhlrc3
|
NHL repeat containing 3 |
| chr19_+_55883924 | 0.01 |
ENSMUST00000111646.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr14_-_72840373 | 0.01 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr7_+_126359763 | 0.01 |
ENSMUST00000091328.4
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr4_-_26346882 | 0.01 |
ENSMUST00000041374.8
ENSMUST00000153813.2 |
Manea
|
mannosidase, endo-alpha |
| chr1_-_45965661 | 0.01 |
ENSMUST00000186804.2
ENSMUST00000187406.7 ENSMUST00000187420.7 |
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr10_-_4337435 | 0.01 |
ENSMUST00000100077.5
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr14_+_102078038 | 0.01 |
ENSMUST00000159314.8
|
Lmo7
|
LIM domain only 7 |
| chr12_-_16850674 | 0.01 |
ENSMUST00000162112.8
|
Greb1
|
gene regulated by estrogen in breast cancer protein |
| chr14_+_61836944 | 0.01 |
ENSMUST00000039562.8
|
Trim13
|
tripartite motif-containing 13 |
| chr8_-_41507808 | 0.01 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr16_-_88759753 | 0.01 |
ENSMUST00000179707.3
|
Krtap16-3
|
keratin associated protein 16-3 |
| chr7_-_84059321 | 0.01 |
ENSMUST00000085077.5
ENSMUST00000207769.2 |
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr6_+_83114086 | 0.01 |
ENSMUST00000087938.11
|
Rtkn
|
rhotekin |
| chr4_+_8691303 | 0.01 |
ENSMUST00000051558.10
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf7l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.4 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.3 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.2 | GO:0032423 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 0.1 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0090327 | cell communication by chemical coupling(GO:0010643) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |