Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
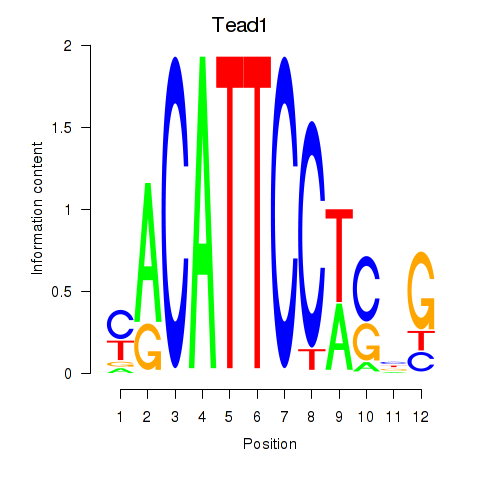
Results for Tead1
Z-value: 1.00
Transcription factors associated with Tead1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tead1
|
ENSMUSG00000055320.19 | TEA domain family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead1 | mm39_v1_chr7_+_112278520_112278532 | -0.80 | 1.1e-01 | Click! |
Activity profile of Tead1 motif
Sorted Z-values of Tead1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_48443525 | 1.40 |
ENSMUST00000057561.9
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2 |
| chr15_+_6416229 | 1.02 |
ENSMUST00000110664.9
ENSMUST00000110663.9 ENSMUST00000161812.8 ENSMUST00000160134.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr7_-_33832640 | 0.73 |
ENSMUST00000038537.9
|
Wtip
|
WT1-interacting protein |
| chr1_+_135693818 | 0.71 |
ENSMUST00000038945.6
|
Phlda3
|
pleckstrin homology like domain, family A, member 3 |
| chr11_-_115903504 | 0.63 |
ENSMUST00000021114.5
|
Galk1
|
galactokinase 1 |
| chr4_-_43558386 | 0.62 |
ENSMUST00000130353.2
|
Tln1
|
talin 1 |
| chr18_+_50186349 | 0.62 |
ENSMUST00000148159.3
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr8_+_129450766 | 0.61 |
ENSMUST00000149116.2
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta) |
| chr5_-_115438971 | 0.56 |
ENSMUST00000112090.2
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr5_+_117501557 | 0.52 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr18_-_35795233 | 0.45 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr1_-_46927230 | 0.45 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr19_-_23425757 | 0.43 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chr19_-_42117420 | 0.40 |
ENSMUST00000161873.2
ENSMUST00000018965.4 |
Avpi1
|
arginine vasopressin-induced 1 |
| chr6_+_146789978 | 0.39 |
ENSMUST00000016631.14
ENSMUST00000203730.3 ENSMUST00000111623.9 |
Ppfibp1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr8_-_106692668 | 0.39 |
ENSMUST00000116429.9
ENSMUST00000034370.17 |
Slc12a4
|
solute carrier family 12, member 4 |
| chr7_+_140796537 | 0.36 |
ENSMUST00000141804.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr15_+_6416079 | 0.35 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr17_-_71309815 | 0.35 |
ENSMUST00000123686.8
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr4_+_152381662 | 0.33 |
ENSMUST00000048892.14
|
Icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr6_-_134864731 | 0.32 |
ENSMUST00000203762.4
ENSMUST00000215088.2 ENSMUST00000066107.10 |
Gpr19
|
G protein-coupled receptor 19 |
| chr18_-_35795175 | 0.32 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr17_+_5991555 | 0.31 |
ENSMUST00000115791.10
ENSMUST00000080283.13 |
Synj2
|
synaptojanin 2 |
| chr4_+_45965327 | 0.29 |
ENSMUST00000107777.9
|
Tdrd7
|
tudor domain containing 7 |
| chr8_+_57964956 | 0.28 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr9_-_62029877 | 0.27 |
ENSMUST00000185675.7
|
Glce
|
glucuronyl C5-epimerase |
| chr5_+_136996713 | 0.26 |
ENSMUST00000001790.6
|
Cldn15
|
claudin 15 |
| chr11_+_68961599 | 0.25 |
ENSMUST00000075980.12
ENSMUST00000094081.5 |
Tmem107
|
transmembrane protein 107 |
| chr7_-_5128936 | 0.24 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chrX_+_100298134 | 0.24 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr4_+_149629559 | 0.23 |
ENSMUST00000105692.2
|
Ctnnbip1
|
catenin beta interacting protein 1 |
| chr7_+_65693350 | 0.23 |
ENSMUST00000206065.2
|
Gm45213
|
predicted gene 45213 |
| chr3_+_96011810 | 0.22 |
ENSMUST00000132980.8
ENSMUST00000138206.8 ENSMUST00000090785.9 ENSMUST00000035519.12 |
Otud7b
|
OTU domain containing 7B |
| chr14_-_54815000 | 0.22 |
ENSMUST00000054487.10
|
Ajuba
|
ajuba LIM protein |
| chr3_+_31049896 | 0.22 |
ENSMUST00000108249.9
|
Prkci
|
protein kinase C, iota |
| chrX_-_103457431 | 0.20 |
ENSMUST00000033695.6
|
Abcb7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr8_-_111808488 | 0.19 |
ENSMUST00000188466.7
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr8_-_111808674 | 0.19 |
ENSMUST00000039597.14
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr2_+_120235239 | 0.18 |
ENSMUST00000135074.2
|
Ganc
|
glucosidase, alpha; neutral C |
| chr2_+_91287846 | 0.18 |
ENSMUST00000028689.4
|
Lrp4
|
low density lipoprotein receptor-related protein 4 |
| chr4_+_15265798 | 0.17 |
ENSMUST00000062684.9
|
Tmem64
|
transmembrane protein 64 |
| chr18_+_50164043 | 0.17 |
ENSMUST00000145726.2
ENSMUST00000128377.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr11_+_70591299 | 0.16 |
ENSMUST00000152618.9
ENSMUST00000102554.8 ENSMUST00000094499.11 ENSMUST00000072187.12 ENSMUST00000137119.3 |
Kif1c
|
kinesin family member 1C |
| chr7_+_130247912 | 0.16 |
ENSMUST00000207549.2
ENSMUST00000209108.2 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_+_189460461 | 0.16 |
ENSMUST00000097442.9
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chrX_+_20714782 | 0.15 |
ENSMUST00000001155.11
ENSMUST00000122312.8 ENSMUST00000120356.8 ENSMUST00000122850.2 |
Araf
|
Araf proto-oncogene, serine/threonine kinase |
| chr7_+_140796096 | 0.14 |
ENSMUST00000153081.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr6_+_88701810 | 0.14 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr14_+_70815250 | 0.14 |
ENSMUST00000228554.2
|
Nudt18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr17_-_36012932 | 0.13 |
ENSMUST00000166980.9
ENSMUST00000145900.8 |
Ddr1
|
discoidin domain receptor family, member 1 |
| chr15_-_36283244 | 0.13 |
ENSMUST00000228358.2
ENSMUST00000022890.10 |
Rnf19a
|
ring finger protein 19A |
| chr2_-_11506511 | 0.13 |
ENSMUST00000183869.8
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr1_+_192984278 | 0.13 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3 |
| chr19_-_57185988 | 0.12 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr2_-_120234539 | 0.12 |
ENSMUST00000090042.12
ENSMUST00000110729.2 ENSMUST00000090046.12 |
Tmem87a
|
transmembrane protein 87A |
| chr11_-_55310724 | 0.10 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr9_-_44624496 | 0.10 |
ENSMUST00000144251.8
ENSMUST00000156918.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr4_-_129636073 | 0.09 |
ENSMUST00000066257.6
|
Khdrbs1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr6_+_88701578 | 0.09 |
ENSMUST00000150180.4
ENSMUST00000163271.8 |
Mgll
|
monoglyceride lipase |
| chr14_-_104705420 | 0.09 |
ENSMUST00000053016.10
|
Pou4f1
|
POU domain, class 4, transcription factor 1 |
| chr8_+_86567600 | 0.09 |
ENSMUST00000053771.14
ENSMUST00000161850.8 |
Phkb
|
phosphorylase kinase beta |
| chr5_+_21850811 | 0.08 |
ENSMUST00000148873.8
ENSMUST00000072896.13 |
Armc10
|
armadillo repeat containing 10 |
| chr8_-_70755132 | 0.08 |
ENSMUST00000008004.10
|
Ddx49
|
DEAD box helicase 49 |
| chr12_+_55349422 | 0.08 |
ENSMUST00000021411.15
|
Prorp
|
protein only RNase P catalytic subunit |
| chr5_+_24631829 | 0.07 |
ENSMUST00000155598.8
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr7_+_140796559 | 0.07 |
ENSMUST00000148975.3
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr17_-_71575584 | 0.07 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr14_-_70585874 | 0.07 |
ENSMUST00000152067.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr11_-_100861713 | 0.06 |
ENSMUST00000060792.6
|
Cavin1
|
caveolae associated 1 |
| chr11_-_85125889 | 0.06 |
ENSMUST00000018625.10
|
Appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr5_+_77099229 | 0.06 |
ENSMUST00000141687.2
|
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr7_+_24603779 | 0.06 |
ENSMUST00000206906.2
ENSMUST00000206011.2 ENSMUST00000117419.8 ENSMUST00000205295.2 |
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr14_-_123150497 | 0.06 |
ENSMUST00000162164.2
ENSMUST00000110679.9 ENSMUST00000161322.3 ENSMUST00000038075.12 |
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr10_+_93476903 | 0.06 |
ENSMUST00000020204.5
|
Ntn4
|
netrin 4 |
| chr1_-_168259710 | 0.06 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr12_+_9023892 | 0.06 |
ENSMUST00000085745.13
ENSMUST00000111113.3 |
Wdr35
|
WD repeat domain 35 |
| chr2_+_90948481 | 0.06 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr18_+_44513547 | 0.05 |
ENSMUST00000202306.2
ENSMUST00000025350.10 |
Dcp2
|
decapping mRNA 2 |
| chr11_-_115824290 | 0.05 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr13_-_34261975 | 0.05 |
ENSMUST00000056427.10
|
Tubb2a
|
tubulin, beta 2A class IIA |
| chr4_+_58285957 | 0.05 |
ENSMUST00000081919.12
ENSMUST00000177951.8 ENSMUST00000098059.10 ENSMUST00000179951.2 ENSMUST00000102893.10 ENSMUST00000084578.12 ENSMUST00000098057.10 |
Musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr11_-_83320281 | 0.04 |
ENSMUST00000052521.9
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr14_+_5894220 | 0.04 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr8_+_46080840 | 0.04 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr19_-_57185861 | 0.04 |
ENSMUST00000111550.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr9_-_8004586 | 0.04 |
ENSMUST00000086580.12
ENSMUST00000065353.13 |
Yap1
|
yes-associated protein 1 |
| chr1_+_4878460 | 0.03 |
ENSMUST00000131119.2
|
Lypla1
|
lysophospholipase 1 |
| chr7_-_142628298 | 0.03 |
ENSMUST00000148715.8
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr1_-_3741721 | 0.03 |
ENSMUST00000070533.5
|
Xkr4
|
X-linked Kx blood group related 4 |
| chr19_-_57185928 | 0.03 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr8_+_57964921 | 0.03 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr10_+_32959472 | 0.03 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr8_+_70755402 | 0.02 |
ENSMUST00000150968.8
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr9_-_65298934 | 0.02 |
ENSMUST00000068307.4
|
Kbtbd13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr17_-_13271183 | 0.02 |
ENSMUST00000091648.4
|
Gpr31b
|
G protein-coupled receptor 31, D17Leh66b region |
| chr9_-_16289527 | 0.02 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr19_+_22670134 | 0.02 |
ENSMUST00000237470.2
ENSMUST00000099564.10 ENSMUST00000099569.10 ENSMUST00000099566.5 ENSMUST00000235712.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr14_-_55204054 | 0.02 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr14_-_55204092 | 0.02 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr14_-_123151162 | 0.02 |
ENSMUST00000160401.8
|
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr11_+_115865535 | 0.02 |
ENSMUST00000021107.14
ENSMUST00000169928.8 ENSMUST00000106461.8 |
Itgb4
|
integrin beta 4 |
| chr6_-_128101275 | 0.02 |
ENSMUST00000127105.8
|
Tspan9
|
tetraspanin 9 |
| chr6_-_98319684 | 0.02 |
ENSMUST00000164491.3
|
Mdfic2
|
MyoD family inhibitor domain containing 2 |
| chr3_-_69767208 | 0.02 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr5_-_86616849 | 0.02 |
ENSMUST00000101073.3
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr19_-_57185808 | 0.02 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr16_+_26281885 | 0.01 |
ENSMUST00000161053.8
ENSMUST00000115302.2 |
Cldn16
|
claudin 16 |
| chr4_+_45203914 | 0.01 |
ENSMUST00000107804.2
|
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr5_-_30401416 | 0.01 |
ENSMUST00000125367.4
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr1_+_135764092 | 0.01 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr2_+_156617329 | 0.01 |
ENSMUST00000088552.7
|
Myl9
|
myosin, light polypeptide 9, regulatory |
| chr8_+_70755168 | 0.01 |
ENSMUST00000066469.14
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr7_+_43645253 | 0.01 |
ENSMUST00000007156.5
|
Klk1b11
|
kallikrein 1-related peptidase b11 |
| chr16_-_89305201 | 0.01 |
ENSMUST00000056118.4
|
Krtap7-1
|
keratin associated protein 7-1 |
| chr10_+_129473353 | 0.01 |
ENSMUST00000214182.2
ENSMUST00000217364.2 ENSMUST00000216446.2 ENSMUST00000213820.2 |
Olfr799
|
olfactory receptor 799 |
| chr15_+_54274151 | 0.01 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr18_-_9450097 | 0.01 |
ENSMUST00000053917.6
|
Ccny
|
cyclin Y |
| chr2_-_63014622 | 0.01 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr3_+_122304778 | 0.01 |
ENSMUST00000198659.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr4_-_118401185 | 0.01 |
ENSMUST00000128098.8
|
Tmem125
|
transmembrane protein 125 |
| chr4_+_137180565 | 0.01 |
ENSMUST00000048893.4
|
1700013G24Rik
|
RIKEN cDNA 1700013G24 gene |
| chr2_+_24181208 | 0.01 |
ENSMUST00000058056.2
|
Il1f10
|
interleukin 1 family, member 10 |
| chr7_+_114344920 | 0.01 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chr18_-_9449947 | 0.01 |
ENSMUST00000234102.2
|
Ccny
|
cyclin Y |
| chr15_+_102875229 | 0.00 |
ENSMUST00000001699.8
|
Hoxc10
|
homeobox C10 |
| chr10_+_24471340 | 0.00 |
ENSMUST00000020171.12
|
Ccn2
|
cellular communication network factor 2 |
| chr16_+_34471558 | 0.00 |
ENSMUST00000023530.5
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr9_-_107486381 | 0.00 |
ENSMUST00000102531.7
ENSMUST00000102530.8 ENSMUST00000195057.2 ENSMUST00000102532.10 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr5_+_103573367 | 0.00 |
ENSMUST00000048957.11
|
Ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr5_+_31274046 | 0.00 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr5_+_31274064 | 0.00 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr11_+_62711057 | 0.00 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr8_+_84267722 | 0.00 |
ENSMUST00000058609.5
|
Olfr370
|
olfactory receptor 370 |
| chr17_-_36013189 | 0.00 |
ENSMUST00000135078.2
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr16_-_34083315 | 0.00 |
ENSMUST00000114953.8
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr11_+_77353431 | 0.00 |
ENSMUST00000130255.2
|
Coro6
|
coronin 6 |
| chr7_-_67913949 | 0.00 |
ENSMUST00000032770.4
ENSMUST00000207874.2 |
Pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr16_-_34083200 | 0.00 |
ENSMUST00000114947.2
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr6_+_87405968 | 0.00 |
ENSMUST00000032125.7
|
Bmp10
|
bone morphogenetic protein 10 |
| chr2_+_82773971 | 0.00 |
ENSMUST00000143764.9
|
Fsip2
|
fibrous sheath-interacting protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tead1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 1.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 0.6 | GO:0006059 | hexitol metabolic process(GO:0006059) glycolytic process from galactose(GO:0061623) |
| 0.2 | 0.6 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.6 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.0 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.0 | 0.2 | GO:0035089 | negative regulation of glial cell apoptotic process(GO:0034351) establishment of apical/basal cell polarity(GO:0035089) Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 0.6 | GO:0034677 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha7-beta1 complex(GO:0034677) integrin alpha10-beta1 complex(GO:0034680) integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 0.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.6 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |