Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Tead3_Tead4
Z-value: 1.25
Transcription factors associated with Tead3_Tead4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
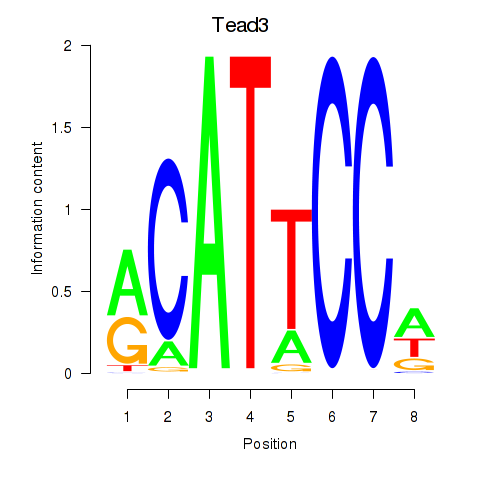
Tead3
|
ENSMUSG00000002249.22 | TEA domain family member 3 |
|
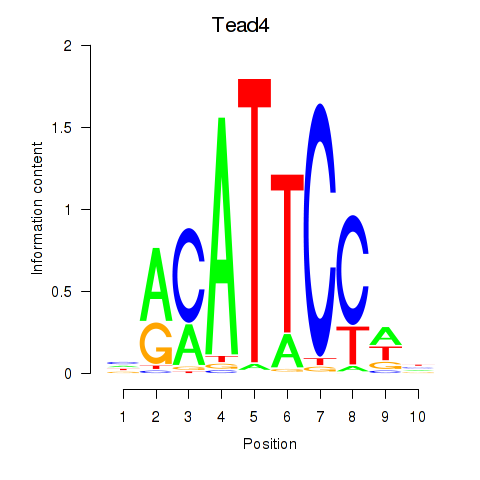
Tead4
|
ENSMUSG00000030353.16 | TEA domain family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead3 | mm39_v1_chr17_-_28569721_28569794 | 0.57 | 3.1e-01 | Click! |
| Tead4 | mm39_v1_chr6_-_128277701_128277786 | 0.33 | 5.9e-01 | Click! |
Activity profile of Tead3_Tead4 motif
Sorted Z-values of Tead3_Tead4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_129854256 | 1.36 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr5_-_66308421 | 0.97 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr2_-_18053158 | 0.89 |
ENSMUST00000066885.6
|
Skida1
|
SKI/DACH domain containing 1 |
| chr5_-_66238313 | 0.87 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr2_-_60711706 | 0.85 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr11_-_69289052 | 0.85 |
ENSMUST00000050140.6
|
Tmem88
|
transmembrane protein 88 |
| chr17_-_71309012 | 0.79 |
ENSMUST00000128179.2
ENSMUST00000150456.2 ENSMUST00000233357.2 ENSMUST00000233417.2 |
Myl12a
Gm49909
|
myosin, light chain 12A, regulatory, non-sarcomeric predicted gene, 49909 |
| chr17_+_47747657 | 0.65 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr7_+_100355910 | 0.64 |
ENSMUST00000207875.2
ENSMUST00000208013.2 |
Fam168a
|
family with sequence similarity 168, member A |
| chr2_+_19662529 | 0.64 |
ENSMUST00000052168.6
|
Otud1
|
OTU domain containing 1 |
| chr12_+_108145802 | 0.59 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr8_-_11728727 | 0.57 |
ENSMUST00000033906.11
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr6_-_99497900 | 0.55 |
ENSMUST00000176565.8
|
Foxp1
|
forkhead box P1 |
| chr15_-_101268036 | 0.54 |
ENSMUST00000077196.6
|
Krt80
|
keratin 80 |
| chr6_-_37419030 | 0.53 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr13_+_112600604 | 0.53 |
ENSMUST00000183663.8
ENSMUST00000184311.8 ENSMUST00000183886.8 |
Il6st
|
interleukin 6 signal transducer |
| chr4_+_132903646 | 0.49 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr6_-_28831746 | 0.48 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr1_-_74040723 | 0.47 |
ENSMUST00000190389.7
|
Tns1
|
tensin 1 |
| chr2_-_18053595 | 0.47 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr2_+_30485048 | 0.46 |
ENSMUST00000102853.4
|
Cstad
|
CSA-conditional, T cell activation-dependent protein |
| chr5_-_66308666 | 0.46 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr12_+_108145997 | 0.45 |
ENSMUST00000101055.5
|
Ccnk
|
cyclin K |
| chr1_+_156386327 | 0.45 |
ENSMUST00000173929.8
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr19_+_8966641 | 0.45 |
ENSMUST00000092956.4
ENSMUST00000092955.11 |
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr4_-_132125510 | 0.42 |
ENSMUST00000136711.2
ENSMUST00000084249.11 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr9_-_87613301 | 0.41 |
ENSMUST00000034991.8
|
Tbx18
|
T-box18 |
| chr14_+_20732804 | 0.41 |
ENSMUST00000228545.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr2_-_90410922 | 0.39 |
ENSMUST00000168621.3
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr1_+_107456731 | 0.39 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr4_+_42949814 | 0.39 |
ENSMUST00000037872.10
ENSMUST00000098112.9 |
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr11_-_115968745 | 0.39 |
ENSMUST00000156545.2
ENSMUST00000075036.9 ENSMUST00000106451.8 |
Unc13d
|
unc-13 homolog D |
| chr11_-_5331734 | 0.39 |
ENSMUST00000172492.8
|
Znrf3
|
zinc and ring finger 3 |
| chr7_+_128125339 | 0.38 |
ENSMUST00000033136.9
|
Bag3
|
BCL2-associated athanogene 3 |
| chr7_-_19432129 | 0.37 |
ENSMUST00000172808.2
ENSMUST00000174191.2 |
Apoe
|
apolipoprotein E |
| chr1_+_55445033 | 0.37 |
ENSMUST00000042986.10
|
Plcl1
|
phospholipase C-like 1 |
| chr12_-_80159768 | 0.37 |
ENSMUST00000219642.2
ENSMUST00000165114.2 ENSMUST00000218835.2 ENSMUST00000021552.3 |
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr19_-_32188413 | 0.37 |
ENSMUST00000151289.9
|
Sgms1
|
sphingomyelin synthase 1 |
| chr5_-_135518098 | 0.37 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr7_-_19432308 | 0.36 |
ENSMUST00000173739.8
|
Apoe
|
apolipoprotein E |
| chr8_-_84963653 | 0.35 |
ENSMUST00000039480.7
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr10_+_53213763 | 0.35 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr12_-_57244096 | 0.34 |
ENSMUST00000044634.12
|
Slc25a21
|
solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 |
| chr4_-_139695337 | 0.34 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr14_-_57983511 | 0.33 |
ENSMUST00000173990.8
ENSMUST00000022531.14 |
Lats2
|
large tumor suppressor 2 |
| chr7_-_44542098 | 0.33 |
ENSMUST00000003049.8
|
Med25
|
mediator complex subunit 25 |
| chr2_+_37406630 | 0.32 |
ENSMUST00000065441.7
|
Gpr21
|
G protein-coupled receptor 21 |
| chr2_+_90865958 | 0.32 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr8_-_123278054 | 0.31 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr6_-_42350188 | 0.31 |
ENSMUST00000073387.5
ENSMUST00000204357.2 |
Epha1
|
Eph receptor A1 |
| chr6_+_37507108 | 0.30 |
ENSMUST00000040987.11
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr6_-_90693471 | 0.30 |
ENSMUST00000101153.10
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr11_+_96740689 | 0.29 |
ENSMUST00000018816.14
|
Copz2
|
coatomer protein complex, subunit zeta 2 |
| chr1_+_138891155 | 0.29 |
ENSMUST00000200533.5
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr5_+_63969706 | 0.29 |
ENSMUST00000081747.8
ENSMUST00000196575.5 |
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chr11_+_116324913 | 0.29 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr3_-_88366159 | 0.29 |
ENSMUST00000147200.8
ENSMUST00000169222.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr7_-_24459736 | 0.29 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr12_+_76580386 | 0.29 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr7_-_29204812 | 0.28 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr15_+_102885467 | 0.27 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9 |
| chr5_+_64969679 | 0.27 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chrX_+_150961960 | 0.27 |
ENSMUST00000096275.5
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr12_+_59142439 | 0.26 |
ENSMUST00000219140.3
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr13_-_95661726 | 0.25 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr8_+_122677231 | 0.25 |
ENSMUST00000093078.13
ENSMUST00000170857.8 ENSMUST00000026354.15 ENSMUST00000174753.8 ENSMUST00000172511.8 |
Banp
|
BTG3 associated nuclear protein |
| chr17_+_29077385 | 0.25 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr2_-_130748259 | 0.25 |
ENSMUST00000128176.2
ENSMUST00000133766.2 ENSMUST00000135110.2 ENSMUST00000146975.2 |
A730017L22Rik
4930402H24Rik
|
RIKEN cDNA A730017L22 gene RIKEN cDNA 4930402H24 gene |
| chr10_-_22696025 | 0.24 |
ENSMUST00000218002.2
ENSMUST00000049930.9 |
Tcf21
|
transcription factor 21 |
| chr15_-_58953838 | 0.23 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr6_-_142453531 | 0.23 |
ENSMUST00000134191.3
ENSMUST00000239397.2 ENSMUST00000239395.2 ENSMUST00000032373.12 |
Ldhb
|
lactate dehydrogenase B |
| chr11_+_77353218 | 0.23 |
ENSMUST00000102493.8
|
Coro6
|
coronin 6 |
| chr13_+_89687915 | 0.23 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr5_+_97145533 | 0.23 |
ENSMUST00000112974.6
ENSMUST00000035635.10 |
Bmp2k
|
BMP2 inducible kinase |
| chr9_+_75682637 | 0.23 |
ENSMUST00000012281.8
|
Bmp5
|
bone morphogenetic protein 5 |
| chr7_-_28071658 | 0.22 |
ENSMUST00000094644.11
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr15_-_98769056 | 0.22 |
ENSMUST00000178486.9
ENSMUST00000023741.16 |
Kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr3_+_20043315 | 0.22 |
ENSMUST00000173779.2
|
Cp
|
ceruloplasmin |
| chr9_-_44352312 | 0.21 |
ENSMUST00000215980.2
ENSMUST00000098837.3 |
Foxr1
|
forkhead box R1 |
| chr7_+_100355798 | 0.21 |
ENSMUST00000107042.9
ENSMUST00000207564.2 ENSMUST00000049053.9 |
Fam168a
|
family with sequence similarity 168, member A |
| chr9_-_55845369 | 0.21 |
ENSMUST00000214747.2
ENSMUST00000217647.2 ENSMUST00000216595.2 |
Scaper
|
S phase cyclin A-associated protein in the ER |
| chr3_-_88366351 | 0.20 |
ENSMUST00000165898.8
ENSMUST00000127436.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr5_+_77414031 | 0.20 |
ENSMUST00000113449.2
|
Rest
|
RE1-silencing transcription factor |
| chr18_+_62086122 | 0.20 |
ENSMUST00000051720.6
ENSMUST00000235860.2 |
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr3_-_103645311 | 0.20 |
ENSMUST00000029440.10
|
Olfml3
|
olfactomedin-like 3 |
| chr9_-_96634874 | 0.19 |
ENSMUST00000152594.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr7_-_109215960 | 0.19 |
ENSMUST00000077909.9
|
Denn2b
|
DENN domain containing 2B |
| chr4_+_134847949 | 0.19 |
ENSMUST00000056977.14
|
Runx3
|
runt related transcription factor 3 |
| chr4_-_119272690 | 0.19 |
ENSMUST00000238287.2
ENSMUST00000238759.2 ENSMUST00000063642.10 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr17_+_47747540 | 0.19 |
ENSMUST00000037701.13
|
AI661453
|
expressed sequence AI661453 |
| chr6_+_125297596 | 0.18 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr11_-_50844572 | 0.18 |
ENSMUST00000162420.2
ENSMUST00000051159.3 |
Prop1
|
paired like homeodomain factor 1 |
| chr9_-_91247809 | 0.17 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr5_-_132570710 | 0.17 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr5_+_135093056 | 0.17 |
ENSMUST00000071263.7
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr5_-_137312424 | 0.17 |
ENSMUST00000199121.2
|
Trip6
|
thyroid hormone receptor interactor 6 |
| chr3_+_94745009 | 0.17 |
ENSMUST00000107266.8
ENSMUST00000042402.12 ENSMUST00000107269.2 |
Pogz
|
pogo transposable element with ZNF domain |
| chr17_+_78815493 | 0.17 |
ENSMUST00000024880.11
ENSMUST00000232859.2 |
Vit
|
vitrin |
| chr15_+_101164719 | 0.17 |
ENSMUST00000230814.2
ENSMUST00000023779.8 |
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr9_+_56982622 | 0.17 |
ENSMUST00000167715.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr18_-_9282754 | 0.16 |
ENSMUST00000041007.4
|
Gjd4
|
gap junction protein, delta 4 |
| chr2_+_164339438 | 0.16 |
ENSMUST00000103101.11
ENSMUST00000117066.8 |
Pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr10_-_127124867 | 0.16 |
ENSMUST00000119078.8
|
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr16_-_16377982 | 0.16 |
ENSMUST00000161861.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_-_87322443 | 0.16 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr7_-_28071919 | 0.16 |
ENSMUST00000119990.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr11_-_115704447 | 0.16 |
ENSMUST00000041684.11
ENSMUST00000156812.2 |
Caskin2
|
CASK-interacting protein 2 |
| chr17_+_3447465 | 0.16 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr9_+_44013019 | 0.16 |
ENSMUST00000034654.9
ENSMUST00000206308.2 ENSMUST00000161381.8 |
Mfrp
|
membrane frizzled-related protein |
| chr2_-_113588983 | 0.16 |
ENSMUST00000099575.4
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr13_-_120252229 | 0.16 |
ENSMUST00000223812.2
|
Zfp131
|
zinc finger protein 131 |
| chr12_+_85793313 | 0.16 |
ENSMUST00000040461.4
|
Flvcr2
|
feline leukemia virus subgroup C cellular receptor 2 |
| chr11_+_112673041 | 0.16 |
ENSMUST00000000579.3
|
Sox9
|
SRY (sex determining region Y)-box 9 |
| chr3_-_95725944 | 0.15 |
ENSMUST00000200164.5
ENSMUST00000090791.8 ENSMUST00000197449.2 |
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr19_-_14575395 | 0.15 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr10_-_61288437 | 0.15 |
ENSMUST00000167087.2
ENSMUST00000020288.15 |
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr19_+_26582450 | 0.15 |
ENSMUST00000176769.9
ENSMUST00000208163.2 ENSMUST00000025862.15 ENSMUST00000176030.8 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_+_12364643 | 0.15 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chr7_-_132415528 | 0.15 |
ENSMUST00000097998.9
|
Fam53b
|
family with sequence similarity 53, member B |
| chr6_-_116084810 | 0.15 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr17_-_47321430 | 0.14 |
ENSMUST00000113337.10
ENSMUST00000113335.4 |
Ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr7_-_65020655 | 0.14 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr17_-_24428351 | 0.14 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr15_+_7120089 | 0.14 |
ENSMUST00000228723.2
|
Lifr
|
LIF receptor alpha |
| chr16_+_20492861 | 0.14 |
ENSMUST00000151679.3
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr11_+_101875095 | 0.14 |
ENSMUST00000176722.8
ENSMUST00000175972.2 |
Cfap97d1
|
CFAP97 domain containing 1 |
| chr8_-_65582206 | 0.14 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr17_-_34846323 | 0.14 |
ENSMUST00000168709.3
ENSMUST00000064953.15 ENSMUST00000170345.8 ENSMUST00000171121.9 ENSMUST00000168391.9 ENSMUST00000169067.9 |
Gm20460
Ppt2
|
predicted gene 20460 palmitoyl-protein thioesterase 2 |
| chr15_+_30172716 | 0.14 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr9_-_15539193 | 0.14 |
ENSMUST00000045513.13
ENSMUST00000152377.2 ENSMUST00000115593.10 |
Deup1
|
deuterosome assembly protein 1 |
| chr15_-_53209513 | 0.13 |
ENSMUST00000077273.9
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr10_+_29087602 | 0.13 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr9_-_103569984 | 0.13 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr10_+_128540049 | 0.13 |
ENSMUST00000217836.2
|
Pmel
|
premelanosome protein |
| chr19_+_5524933 | 0.13 |
ENSMUST00000236518.2
|
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr4_-_24800890 | 0.13 |
ENSMUST00000108214.9
|
Klhl32
|
kelch-like 32 |
| chr1_-_155848917 | 0.13 |
ENSMUST00000138762.8
|
Cep350
|
centrosomal protein 350 |
| chr16_+_38182569 | 0.13 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr10_-_12702674 | 0.13 |
ENSMUST00000219130.2
|
Utrn
|
utrophin |
| chr1_+_104696235 | 0.13 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr17_-_34846122 | 0.13 |
ENSMUST00000171376.8
ENSMUST00000169287.2 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr8_+_45960804 | 0.12 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr5_-_140687995 | 0.12 |
ENSMUST00000135028.5
ENSMUST00000077890.12 ENSMUST00000041783.14 ENSMUST00000142081.6 |
Iqce
|
IQ motif containing E |
| chr3_-_101017594 | 0.12 |
ENSMUST00000102694.4
|
Ptgfrn
|
prostaglandin F2 receptor negative regulator |
| chr2_-_73316053 | 0.12 |
ENSMUST00000102680.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr5_+_150183201 | 0.12 |
ENSMUST00000087204.9
|
Fry
|
FRY microtubule binding protein |
| chr8_-_95644639 | 0.12 |
ENSMUST00000077955.6
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr18_-_63825380 | 0.12 |
ENSMUST00000025476.4
|
Txnl1
|
thioredoxin-like 1 |
| chr8_-_37081091 | 0.12 |
ENSMUST00000033923.14
|
Dlc1
|
deleted in liver cancer 1 |
| chr5_+_77413282 | 0.12 |
ENSMUST00000080359.12
|
Rest
|
RE1-silencing transcription factor |
| chr2_+_112097087 | 0.12 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr17_-_36012830 | 0.12 |
ENSMUST00000155628.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr6_-_134543876 | 0.12 |
ENSMUST00000032322.15
ENSMUST00000126836.4 |
Lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr3_+_94744844 | 0.11 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr14_-_39194782 | 0.11 |
ENSMUST00000168810.9
ENSMUST00000173780.2 ENSMUST00000166968.9 |
Nrg3
|
neuregulin 3 |
| chr11_+_115824108 | 0.11 |
ENSMUST00000140991.2
|
Sap30bp
|
SAP30 binding protein |
| chr17_-_48002328 | 0.11 |
ENSMUST00000152214.4
ENSMUST00000113299.6 |
Prickle4
|
prickle planar cell polarity protein 4 |
| chr2_-_69619864 | 0.11 |
ENSMUST00000094942.4
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr5_-_121975676 | 0.11 |
ENSMUST00000197892.3
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr18_-_32595515 | 0.11 |
ENSMUST00000207669.2
|
Gm35060
|
predicted gene, 35060 |
| chr10_+_126814542 | 0.11 |
ENSMUST00000105256.10
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr19_-_59334172 | 0.11 |
ENSMUST00000099274.4
|
Pdzd8
|
PDZ domain containing 8 |
| chr19_+_46140942 | 0.10 |
ENSMUST00000026254.14
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr16_+_32151056 | 0.10 |
ENSMUST00000115151.5
ENSMUST00000232137.2 |
Ubxn7
|
UBX domain protein 7 |
| chr11_-_115968576 | 0.10 |
ENSMUST00000106450.8
|
Unc13d
|
unc-13 homolog D |
| chr19_+_42034231 | 0.10 |
ENSMUST00000172244.8
ENSMUST00000081714.5 |
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr6_+_113435716 | 0.10 |
ENSMUST00000203661.3
ENSMUST00000204774.3 ENSMUST00000053569.7 ENSMUST00000101065.8 |
Il17re
|
interleukin 17 receptor E |
| chr11_+_98303287 | 0.10 |
ENSMUST00000058295.6
|
Erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr8_-_105350898 | 0.10 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr7_-_109215754 | 0.09 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chr17_+_75772475 | 0.09 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr2_-_39080701 | 0.09 |
ENSMUST00000142872.2
ENSMUST00000038874.12 |
Scai
|
suppressor of cancer cell invasion |
| chr9_+_27210500 | 0.09 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr17_+_47904355 | 0.09 |
ENSMUST00000182209.8
|
Ccnd3
|
cyclin D3 |
| chr2_+_72115981 | 0.09 |
ENSMUST00000090824.12
ENSMUST00000135469.8 |
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr4_-_4138432 | 0.09 |
ENSMUST00000070375.8
|
Penk
|
preproenkephalin |
| chr6_+_121277186 | 0.09 |
ENSMUST00000064580.14
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr6_-_120341304 | 0.08 |
ENSMUST00000146667.2
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr10_+_27950809 | 0.08 |
ENSMUST00000166468.2
ENSMUST00000218359.2 ENSMUST00000218276.2 |
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr9_-_58943670 | 0.08 |
ENSMUST00000214547.2
ENSMUST00000068664.7 |
Neo1
|
neogenin |
| chr10_-_42152684 | 0.08 |
ENSMUST00000175881.8
ENSMUST00000056974.4 ENSMUST00000105502.8 ENSMUST00000105501.2 |
Foxo3
|
forkhead box O3 |
| chr9_+_108924457 | 0.08 |
ENSMUST00000072093.13
|
Plxnb1
|
plexin B1 |
| chr11_-_60804601 | 0.08 |
ENSMUST00000019075.4
|
Natd1
|
N-acetyltransferase domain containing 1 |
| chr2_-_165700055 | 0.08 |
ENSMUST00000109266.11
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr2_-_148285450 | 0.08 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr14_-_103336990 | 0.08 |
ENSMUST00000022720.15
ENSMUST00000144141.8 |
Fbxl3
|
F-box and leucine-rich repeat protein 3 |
| chr13_+_109769294 | 0.08 |
ENSMUST00000135275.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr11_-_115698894 | 0.08 |
ENSMUST00000132780.8
|
Caskin2
|
CASK-interacting protein 2 |
| chr10_-_78427721 | 0.08 |
ENSMUST00000040580.7
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr3_+_90561560 | 0.08 |
ENSMUST00000079286.4
|
S100a7a
|
S100 calcium binding protein A7A |
| chr6_+_17306334 | 0.08 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr18_+_82572595 | 0.08 |
ENSMUST00000152071.9
ENSMUST00000142850.9 ENSMUST00000080658.12 ENSMUST00000133193.9 ENSMUST00000123251.9 ENSMUST00000153478.9 ENSMUST00000075372.13 ENSMUST00000102812.12 ENSMUST00000062446.15 ENSMUST00000114674.11 ENSMUST00000132369.3 |
Mbp
|
myelin basic protein |
| chr10_+_129263175 | 0.08 |
ENSMUST00000204529.3
|
Olfr786
|
olfactory receptor 786 |
| chr1_+_106099482 | 0.08 |
ENSMUST00000061047.7
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr7_-_28072022 | 0.07 |
ENSMUST00000144700.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr4_+_57568144 | 0.07 |
ENSMUST00000102904.10
|
Pakap
|
paralemmin A kinase anchor protein |
| chr17_-_28569574 | 0.07 |
ENSMUST00000114799.8
ENSMUST00000219703.3 |
Tead3
|
TEA domain family member 3 |
| chr16_+_43184191 | 0.07 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr15_+_101308935 | 0.07 |
ENSMUST00000147662.8
|
Krt7
|
keratin 7 |
| chr6_-_50573604 | 0.07 |
ENSMUST00000031852.5
|
4921507P07Rik
|
RIKEN cDNA 4921507P07 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tead3_Tead4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.2 | 0.8 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 2.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.5 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 | 0.4 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 1.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.5 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.1 | 0.3 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.4 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.3 | GO:0070949 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.3 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 0.2 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.5 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.2 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) renal vesicle induction(GO:0072034) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 1.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.4 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0061347 | planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.2 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.4 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.0 | 0.1 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.0 | 0.2 | GO:1901626 | regulation of postsynaptic membrane organization(GO:1901626) |
| 0.0 | 0.2 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.0 | 0.1 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0035926 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.0 | 0.3 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.5 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.5 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.0 | 0.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.0 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.2 | 0.5 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.7 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.3 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.2 | 0.5 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 1.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |