Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
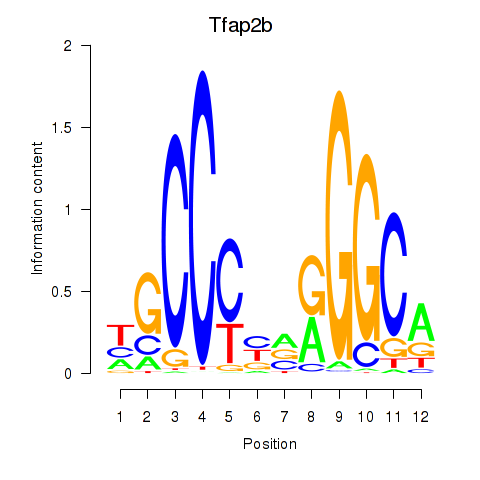
Results for Tfap2b
Z-value: 0.51
Transcription factors associated with Tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2b
|
ENSMUSG00000025927.14 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2b | mm39_v1_chr1_+_19279178_19279191 | 0.93 | 2.3e-02 | Click! |
Activity profile of Tfap2b motif
Sorted Z-values of Tfap2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_76756772 | 0.62 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr15_-_98769056 | 0.40 |
ENSMUST00000178486.9
ENSMUST00000023741.16 |
Kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr11_-_102255999 | 0.39 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr1_-_63153414 | 0.33 |
ENSMUST00000153992.2
ENSMUST00000165066.8 ENSMUST00000172416.8 ENSMUST00000137511.8 |
Ino80d
|
INO80 complex subunit D |
| chr4_-_57143437 | 0.31 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr9_+_118881838 | 0.30 |
ENSMUST00000051386.13
ENSMUST00000074734.13 |
Vill
|
villin-like |
| chr2_-_5947551 | 0.29 |
ENSMUST00000026924.7
ENSMUST00000095147.9 |
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr10_-_59787646 | 0.27 |
ENSMUST00000020308.5
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chr11_-_70146156 | 0.26 |
ENSMUST00000108574.3
ENSMUST00000000329.9 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr7_-_127048280 | 0.25 |
ENSMUST00000053392.11
|
Zfp689
|
zinc finger protein 689 |
| chr3_-_31023547 | 0.24 |
ENSMUST00000064718.12
ENSMUST00000177992.8 ENSMUST00000129817.9 ENSMUST00000168645.8 ENSMUST00000108255.8 ENSMUST00000099163.5 |
Phc3
|
polyhomeotic 3 |
| chr11_-_116718857 | 0.23 |
ENSMUST00000021170.9
|
Mxra7
|
matrix-remodelling associated 7 |
| chr17_-_32503689 | 0.23 |
ENSMUST00000127893.8
|
Brd4
|
bromodomain containing 4 |
| chr7_+_24990596 | 0.23 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr13_+_55097200 | 0.22 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr7_-_97958679 | 0.22 |
ENSMUST00000033020.14
|
Acer3
|
alkaline ceramidase 3 |
| chr18_+_61058684 | 0.22 |
ENSMUST00000102888.10
ENSMUST00000025519.11 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr15_-_76116754 | 0.21 |
ENSMUST00000166428.8
|
Plec
|
plectin |
| chr17_-_32503107 | 0.20 |
ENSMUST00000237692.2
|
Brd4
|
bromodomain containing 4 |
| chr15_-_76116245 | 0.20 |
ENSMUST00000167754.8
|
Plec
|
plectin |
| chr18_+_82929451 | 0.20 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr2_+_119004964 | 0.20 |
ENSMUST00000239130.2
ENSMUST00000069711.3 |
Gm14137
|
predicted gene 14137 |
| chr8_-_123768984 | 0.19 |
ENSMUST00000212937.2
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr7_+_28834276 | 0.19 |
ENSMUST00000161522.8
ENSMUST00000204845.3 ENSMUST00000205027.3 ENSMUST00000204194.3 ENSMUST00000203070.3 ENSMUST00000203380.3 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr12_+_76580386 | 0.18 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr7_+_28834350 | 0.17 |
ENSMUST00000159975.8
ENSMUST00000094617.11 ENSMUST00000032811.12 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr16_+_58490625 | 0.16 |
ENSMUST00000060077.7
|
Cpox
|
coproporphyrinogen oxidase |
| chr2_+_30127692 | 0.16 |
ENSMUST00000113654.8
ENSMUST00000095078.3 |
Lrrc8a
|
leucine rich repeat containing 8A VRAC subunit A |
| chr6_-_97464761 | 0.16 |
ENSMUST00000032146.14
|
Frmd4b
|
FERM domain containing 4B |
| chr7_+_127078371 | 0.16 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chr4_+_8690398 | 0.15 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr13_+_41071077 | 0.15 |
ENSMUST00000067778.8
ENSMUST00000225759.2 |
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr11_+_32483290 | 0.15 |
ENSMUST00000102821.4
|
Stk10
|
serine/threonine kinase 10 |
| chr7_+_86645323 | 0.15 |
ENSMUST00000233714.2
ENSMUST00000233648.2 ENSMUST00000164462.3 ENSMUST00000233730.2 |
Vmn2r79
|
vomeronasal 2, receptor 79 |
| chr4_+_132903646 | 0.15 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr7_+_28834391 | 0.15 |
ENSMUST00000160194.8
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr9_-_52079872 | 0.14 |
ENSMUST00000213645.2
|
Zc3h12c
|
zinc finger CCCH type containing 12C |
| chr18_+_36693024 | 0.14 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr16_+_24266829 | 0.14 |
ENSMUST00000078988.10
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr15_-_103242697 | 0.14 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr12_-_111638722 | 0.14 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr7_+_46045862 | 0.14 |
ENSMUST00000025202.8
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr1_-_96799832 | 0.14 |
ENSMUST00000071985.6
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr8_+_85449632 | 0.14 |
ENSMUST00000098571.5
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr6_+_113370023 | 0.13 |
ENSMUST00000203524.3
|
Ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr8_-_123404811 | 0.13 |
ENSMUST00000006525.14
ENSMUST00000064674.13 |
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chrX_+_7439839 | 0.13 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr6_-_119825054 | 0.13 |
ENSMUST00000079582.5
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr14_-_30075424 | 0.13 |
ENSMUST00000224198.3
ENSMUST00000238675.2 ENSMUST00000112249.10 ENSMUST00000224785.3 |
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr17_+_36152383 | 0.13 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr10_-_67972401 | 0.12 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr1_+_131671751 | 0.12 |
ENSMUST00000049027.10
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr1_+_55445033 | 0.12 |
ENSMUST00000042986.10
|
Plcl1
|
phospholipase C-like 1 |
| chr3_+_90201388 | 0.12 |
ENSMUST00000199607.5
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr15_-_71906051 | 0.12 |
ENSMUST00000159993.8
|
Col22a1
|
collagen, type XXII, alpha 1 |
| chr11_+_107438751 | 0.12 |
ENSMUST00000100305.8
ENSMUST00000075012.8 ENSMUST00000106746.8 |
Helz
|
helicase with zinc finger domain |
| chr5_+_129937377 | 0.12 |
ENSMUST00000178355.3
|
Nupr1l
|
nuclear protein transcriptional regulator 1 like |
| chr8_-_123405392 | 0.12 |
ENSMUST00000134045.2
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr11_-_50183129 | 0.12 |
ENSMUST00000059458.5
|
Maml1
|
mastermind like transcriptional coactivator 1 |
| chr7_+_27879650 | 0.12 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr9_+_37400577 | 0.11 |
ENSMUST00000211060.2
ENSMUST00000239463.2 ENSMUST00000048604.8 |
Msantd2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr4_-_136563154 | 0.11 |
ENSMUST00000105846.9
ENSMUST00000059287.14 ENSMUST00000105845.9 |
Ephb2
|
Eph receptor B2 |
| chr6_+_124986224 | 0.11 |
ENSMUST00000112427.8
|
Zfp384
|
zinc finger protein 384 |
| chr13_+_58956495 | 0.11 |
ENSMUST00000225950.2
ENSMUST00000225583.2 |
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr8_+_106024294 | 0.11 |
ENSMUST00000015003.10
|
E2f4
|
E2F transcription factor 4 |
| chr11_-_115977755 | 0.11 |
ENSMUST00000074628.13
ENSMUST00000106444.4 |
Wbp2
|
WW domain binding protein 2 |
| chr13_+_58956077 | 0.11 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr15_-_38300937 | 0.11 |
ENSMUST00000227920.2
ENSMUST00000074043.7 ENSMUST00000228416.2 |
Klf10
|
Kruppel-like factor 10 |
| chr2_+_158509039 | 0.11 |
ENSMUST00000045503.11
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr17_-_10538253 | 0.10 |
ENSMUST00000233828.2
ENSMUST00000233645.2 ENSMUST00000042296.9 |
Qk
|
quaking, KH domain containing RNA binding |
| chr13_-_103470937 | 0.10 |
ENSMUST00000167058.8
ENSMUST00000164111.2 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_-_91854844 | 0.10 |
ENSMUST00000028663.5
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr9_-_44632680 | 0.10 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr4_+_43669266 | 0.10 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr15_-_102165884 | 0.10 |
ENSMUST00000043172.15
|
Rarg
|
retinoic acid receptor, gamma |
| chr7_+_121666388 | 0.09 |
ENSMUST00000033158.6
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr10_-_61288437 | 0.09 |
ENSMUST00000167087.2
ENSMUST00000020288.15 |
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr3_+_152102292 | 0.09 |
ENSMUST00000089982.11
ENSMUST00000106101.2 |
Zzz3
|
zinc finger, ZZ domain containing 3 |
| chr3_-_84387700 | 0.09 |
ENSMUST00000194027.2
ENSMUST00000107689.7 |
Fhdc1
|
FH2 domain containing 1 |
| chr7_+_24981604 | 0.09 |
ENSMUST00000163320.8
ENSMUST00000005578.13 |
Cic
|
capicua transcriptional repressor |
| chrX_-_7440480 | 0.09 |
ENSMUST00000115742.9
ENSMUST00000150787.8 |
Ppp1r3f
|
protein phosphatase 1, regulatory subunit 3F |
| chr6_-_39702127 | 0.09 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr5_-_25703700 | 0.09 |
ENSMUST00000173073.8
ENSMUST00000045291.14 ENSMUST00000173174.2 |
Kmt2c
|
lysine (K)-specific methyltransferase 2C |
| chr1_-_132294807 | 0.09 |
ENSMUST00000136828.3
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr9_-_40366966 | 0.09 |
ENSMUST00000165104.8
ENSMUST00000045682.7 |
Gramd1b
|
GRAM domain containing 1B |
| chr10_-_81037878 | 0.09 |
ENSMUST00000005069.8
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr3_-_113423774 | 0.09 |
ENSMUST00000092154.10
ENSMUST00000106536.8 ENSMUST00000106535.2 |
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chrX_+_158410229 | 0.09 |
ENSMUST00000112456.9
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_-_106504196 | 0.09 |
ENSMUST00000128933.2
|
Tex2
|
testis expressed gene 2 |
| chr16_-_20245138 | 0.08 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr7_-_45045097 | 0.08 |
ENSMUST00000211121.2
ENSMUST00000074575.11 |
Snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr5_-_137312424 | 0.08 |
ENSMUST00000199121.2
|
Trip6
|
thyroid hormone receptor interactor 6 |
| chr2_+_30176418 | 0.08 |
ENSMUST00000138666.8
ENSMUST00000113634.3 |
Nup188
|
nucleoporin 188 |
| chr2_-_91014163 | 0.08 |
ENSMUST00000077941.13
ENSMUST00000111381.9 ENSMUST00000111372.8 ENSMUST00000111371.8 ENSMUST00000075269.10 ENSMUST00000066473.12 |
Madd
|
MAP-kinase activating death domain |
| chr14_-_55909314 | 0.08 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr15_-_11905697 | 0.08 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr3_+_141170941 | 0.08 |
ENSMUST00000106236.9
|
Unc5c
|
unc-5 netrin receptor C |
| chr9_+_21095399 | 0.07 |
ENSMUST00000115458.9
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr7_+_79882609 | 0.07 |
ENSMUST00000062915.9
|
Gdpgp1
|
GDP-D-glucose phosphorylase 1 |
| chr16_-_20245071 | 0.07 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_-_13444249 | 0.07 |
ENSMUST00000068304.13
ENSMUST00000006037.13 |
Ncoa2
|
nuclear receptor coactivator 2 |
| chr3_-_137687284 | 0.07 |
ENSMUST00000136613.4
ENSMUST00000029806.13 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chrX_-_9335525 | 0.07 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr5_+_149335214 | 0.07 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr9_+_61280890 | 0.07 |
ENSMUST00000161689.8
|
Tle3
|
transducin-like enhancer of split 3 |
| chr2_-_63928339 | 0.07 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr16_+_3561952 | 0.07 |
ENSMUST00000023176.5
|
Zfp263
|
zinc finger protein 263 |
| chr6_-_118539187 | 0.07 |
ENSMUST00000112830.3
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr19_+_6413703 | 0.07 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr1_-_57011595 | 0.07 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr10_+_19232281 | 0.07 |
ENSMUST00000053225.7
|
Olig3
|
oligodendrocyte transcription factor 3 |
| chr2_+_158508609 | 0.06 |
ENSMUST00000103116.10
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr8_+_117983803 | 0.06 |
ENSMUST00000166750.9
|
Cmip
|
c-Maf inducing protein |
| chr1_+_86082474 | 0.06 |
ENSMUST00000113309.9
ENSMUST00000186509.7 ENSMUST00000027434.15 ENSMUST00000131412.8 |
Armc9
|
armadillo repeat containing 9 |
| chrX_+_9138995 | 0.06 |
ENSMUST00000015486.7
|
Xk
|
X-linked Kx blood group |
| chr3_+_130904000 | 0.06 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr11_-_94133527 | 0.06 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr14_+_45588632 | 0.06 |
ENSMUST00000228818.2
|
Styx
|
serine/threonine/tyrosine interaction protein |
| chr8_-_96058383 | 0.06 |
ENSMUST00000057717.8
|
Zfp319
|
zinc finger protein 319 |
| chr11_-_75345482 | 0.06 |
ENSMUST00000173320.8
|
Wdr81
|
WD repeat domain 81 |
| chr17_-_48189815 | 0.06 |
ENSMUST00000154108.2
|
Foxp4
|
forkhead box P4 |
| chr8_+_106331866 | 0.06 |
ENSMUST00000043531.10
|
Ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr10_+_7556948 | 0.06 |
ENSMUST00000165952.9
|
Lats1
|
large tumor suppressor |
| chr10_-_116984386 | 0.06 |
ENSMUST00000020381.5
|
Frs2
|
fibroblast growth factor receptor substrate 2 |
| chr10_-_81179100 | 0.06 |
ENSMUST00000045102.7
|
Gipc3
|
GIPC PDZ domain containing family, member 3 |
| chr14_-_55909527 | 0.06 |
ENSMUST00000010520.10
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr2_-_35995283 | 0.05 |
ENSMUST00000112960.8
ENSMUST00000112967.12 ENSMUST00000112963.8 |
Lhx6
|
LIM homeobox protein 6 |
| chr16_-_38533597 | 0.05 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr1_-_63153675 | 0.05 |
ENSMUST00000097718.9
|
Ino80d
|
INO80 complex subunit D |
| chr10_-_121312212 | 0.05 |
ENSMUST00000026902.9
|
Rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr9_+_44394080 | 0.05 |
ENSMUST00000220303.2
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr17_+_43327412 | 0.05 |
ENSMUST00000024708.6
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr2_+_155798457 | 0.05 |
ENSMUST00000109619.9
ENSMUST00000094421.11 ENSMUST00000039994.14 ENSMUST00000151569.8 ENSMUST00000109618.2 |
Cep250
|
centrosomal protein 250 |
| chr15_-_102165740 | 0.05 |
ENSMUST00000135466.2
|
Rarg
|
retinoic acid receptor, gamma |
| chr1_-_135032972 | 0.05 |
ENSMUST00000044828.14
|
Lgr6
|
leucine-rich repeat-containing G protein-coupled receptor 6 |
| chr17_+_33774681 | 0.05 |
ENSMUST00000087605.13
ENSMUST00000174695.2 |
Myo1f
|
myosin IF |
| chr15_-_97729341 | 0.05 |
ENSMUST00000079838.14
ENSMUST00000118294.8 |
Hdac7
|
histone deacetylase 7 |
| chr12_+_102094977 | 0.05 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chrX_+_72800675 | 0.05 |
ENSMUST00000002079.7
|
Plxnb3
|
plexin B3 |
| chr16_-_20244631 | 0.04 |
ENSMUST00000077867.10
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr2_+_74528071 | 0.04 |
ENSMUST00000059272.10
|
Hoxd9
|
homeobox D9 |
| chr2_+_104716661 | 0.04 |
ENSMUST00000111114.8
|
Ccdc73
|
coiled-coil domain containing 73 |
| chr14_+_61410397 | 0.04 |
ENSMUST00000121091.2
|
Sacs
|
sacsin |
| chr1_+_34840785 | 0.04 |
ENSMUST00000047664.16
ENSMUST00000211073.2 |
Arhgef4
SMIM39
|
Rho guanine nucleotide exchange factor (GEF) 4 novel protein |
| chr1_+_39940043 | 0.04 |
ENSMUST00000168431.7
ENSMUST00000163854.9 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr8_+_79754980 | 0.04 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr7_+_141027557 | 0.04 |
ENSMUST00000106004.8
|
Rplp2
|
ribosomal protein, large P2 |
| chr2_+_148237258 | 0.04 |
ENSMUST00000109962.4
|
Sstr4
|
somatostatin receptor 4 |
| chr2_+_102488985 | 0.04 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr19_+_5790918 | 0.04 |
ENSMUST00000081496.6
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr15_-_75620060 | 0.04 |
ENSMUST00000062002.6
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr14_+_45588509 | 0.04 |
ENSMUST00000226873.2
|
Styx
|
serine/threonine/tyrosine interaction protein |
| chr19_-_3332955 | 0.04 |
ENSMUST00000119292.8
ENSMUST00000025751.11 |
Ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr17_+_34808772 | 0.04 |
ENSMUST00000038244.15
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr6_+_54794433 | 0.04 |
ENSMUST00000127331.2
|
Znrf2
|
zinc and ring finger 2 |
| chr9_-_96993169 | 0.04 |
ENSMUST00000085206.11
|
Slc25a36
|
solute carrier family 25, member 36 |
| chr7_-_66339319 | 0.03 |
ENSMUST00000124899.8
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr2_+_130248398 | 0.03 |
ENSMUST00000055421.6
|
Tmem239
|
transmembrane 239 |
| chr7_-_80020830 | 0.03 |
ENSMUST00000205436.2
ENSMUST00000098346.5 |
Man2a2
|
mannosidase 2, alpha 2 |
| chr11_+_121150798 | 0.03 |
ENSMUST00000106113.2
|
Foxk2
|
forkhead box K2 |
| chr7_+_25386418 | 0.03 |
ENSMUST00000002678.10
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr7_+_58307930 | 0.03 |
ENSMUST00000168747.3
|
Atp10a
|
ATPase, class V, type 10A |
| chr9_+_38288382 | 0.03 |
ENSMUST00000214865.2
|
Olfr251
|
olfactory receptor 251 |
| chr2_-_154411765 | 0.03 |
ENSMUST00000103145.11
|
E2f1
|
E2F transcription factor 1 |
| chr8_-_85663976 | 0.03 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr10_+_81411333 | 0.03 |
ENSMUST00000143285.8
ENSMUST00000146358.8 |
Tle2
|
transducin-like enhancer of split 2 |
| chr2_+_3714515 | 0.03 |
ENSMUST00000115053.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr7_-_97811525 | 0.03 |
ENSMUST00000107112.2
|
Capn5
|
calpain 5 |
| chr17_-_46593917 | 0.03 |
ENSMUST00000225359.2
ENSMUST00000225080.2 ENSMUST00000225943.2 ENSMUST00000012440.14 ENSMUST00000164342.10 ENSMUST00000180283.2 |
Tjap1
|
tight junction associated protein 1 |
| chr6_+_54793894 | 0.03 |
ENSMUST00000079869.13
|
Znrf2
|
zinc and ring finger 2 |
| chr2_-_181335518 | 0.03 |
ENSMUST00000108776.8
ENSMUST00000108771.2 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr11_-_95037089 | 0.03 |
ENSMUST00000021241.8
|
Dlx4
|
distal-less homeobox 4 |
| chr7_-_28001624 | 0.03 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr17_+_29020064 | 0.03 |
ENSMUST00000004985.11
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr18_+_65713827 | 0.02 |
ENSMUST00000182852.3
ENSMUST00000236633.2 ENSMUST00000182319.9 ENSMUST00000182140.8 |
Zfp532
|
zinc finger protein 532 |
| chr2_+_30176395 | 0.02 |
ENSMUST00000064447.12
|
Nup188
|
nucleoporin 188 |
| chr11_-_66416621 | 0.02 |
ENSMUST00000123454.8
|
Shisa6
|
shisa family member 6 |
| chr17_-_46593574 | 0.02 |
ENSMUST00000224230.2
ENSMUST00000225288.2 |
Tjap1
|
tight junction associated protein 1 |
| chr11_+_82671934 | 0.02 |
ENSMUST00000092849.12
ENSMUST00000021039.12 ENSMUST00000080461.12 ENSMUST00000173347.8 ENSMUST00000173727.8 ENSMUST00000173009.8 ENSMUST00000131537.9 ENSMUST00000173722.8 |
Lig3
|
ligase III, DNA, ATP-dependent |
| chr10_-_127047396 | 0.02 |
ENSMUST00000013970.9
|
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr19_+_5100815 | 0.02 |
ENSMUST00000224178.2
ENSMUST00000225799.3 ENSMUST00000025818.8 |
Rin1
|
Ras and Rab interactor 1 |
| chr13_+_55359866 | 0.02 |
ENSMUST00000224693.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr15_+_103411461 | 0.02 |
ENSMUST00000023132.5
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr18_+_61044830 | 0.02 |
ENSMUST00000040359.6
|
Arsi
|
arylsulfatase i |
| chr9_-_44231526 | 0.02 |
ENSMUST00000214602.2
ENSMUST00000065080.10 |
C2cd2l
|
C2 calcium-dependent domain containing 2-like |
| chr1_+_191638854 | 0.02 |
ENSMUST00000044954.7
|
Slc30a1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr8_-_106670014 | 0.02 |
ENSMUST00000038896.8
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr7_-_43896112 | 0.02 |
ENSMUST00000237383.2
|
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr14_+_56122404 | 0.02 |
ENSMUST00000022831.5
|
Khnyn
|
KH and NYN domain containing |
| chr1_-_14380418 | 0.02 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_-_102903680 | 0.01 |
ENSMUST00000132449.8
ENSMUST00000111183.2 ENSMUST00000011058.9 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr11_-_101061153 | 0.01 |
ENSMUST00000123864.2
|
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr15_+_102927366 | 0.01 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr3_+_40663285 | 0.01 |
ENSMUST00000091184.9
|
Slc25a31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr5_+_110378841 | 0.01 |
ENSMUST00000197188.5
ENSMUST00000086674.10 ENSMUST00000031474.11 |
Ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr9_+_35819708 | 0.01 |
ENSMUST00000176049.2
ENSMUST00000176153.2 |
Pate13
|
prostate and testis expressed 13 |
| chr6_-_37276885 | 0.01 |
ENSMUST00000101532.10
|
Dgki
|
diacylglycerol kinase, iota |
| chr17_-_36501112 | 0.01 |
ENSMUST00000025312.13
ENSMUST00000102675.10 |
H2-T3
|
histocompatibility 2, T region locus 3 |
| chr6_+_113435716 | 0.01 |
ENSMUST00000203661.3
ENSMUST00000204774.3 ENSMUST00000053569.7 ENSMUST00000101065.8 |
Il17re
|
interleukin 17 receptor E |
| chr1_-_14380327 | 0.01 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0002654 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.2 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.0 | GO:0002362 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.5 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:2001107 | negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.0 | 0.2 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.2 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.0 | GO:1902462 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.2 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 1.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |