Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
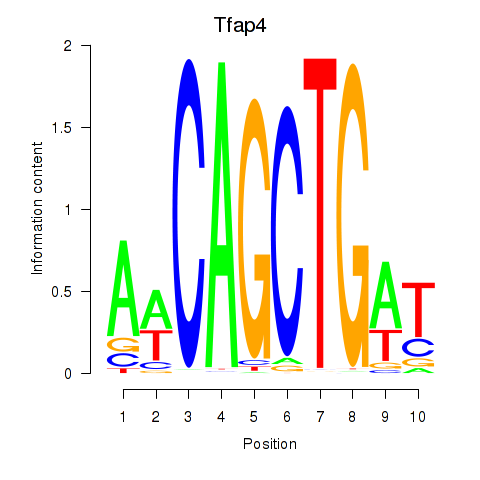
Results for Tfap4
Z-value: 1.07
Transcription factors associated with Tfap4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap4
|
ENSMUSG00000005718.9 | transcription factor AP4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap4 | mm39_v1_chr16_-_4376471_4376647 | 0.68 | 2.1e-01 | Click! |
Activity profile of Tfap4 motif
Sorted Z-values of Tfap4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_32967937 | 0.75 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr8_+_83891972 | 0.69 |
ENSMUST00000034145.11
|
Tbc1d9
|
TBC1 domain family, member 9 |
| chr5_+_90907207 | 0.66 |
ENSMUST00000031318.6
|
Cxcl5
|
chemokine (C-X-C motif) ligand 5 |
| chr15_-_90563510 | 0.65 |
ENSMUST00000014777.9
ENSMUST00000064391.12 |
Cpne8
|
copine VIII |
| chr1_-_16163506 | 0.60 |
ENSMUST00000145070.8
ENSMUST00000151004.2 |
4930444P10Rik
|
RIKEN cDNA 4930444P10 gene |
| chr4_+_119671688 | 0.59 |
ENSMUST00000106307.9
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr8_-_71257623 | 0.56 |
ENSMUST00000212875.2
ENSMUST00000212001.2 ENSMUST00000212757.2 |
Mast3
|
microtubule associated serine/threonine kinase 3 |
| chr1_+_159351337 | 0.56 |
ENSMUST00000192069.6
|
Tnr
|
tenascin R |
| chr13_-_103233323 | 0.55 |
ENSMUST00000166336.9
ENSMUST00000239261.2 ENSMUST00000194446.7 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr18_-_60724855 | 0.53 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr5_+_107655487 | 0.51 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr16_+_14523696 | 0.48 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr4_-_19922599 | 0.48 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr8_-_106198112 | 0.45 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr4_-_139037957 | 0.43 |
ENSMUST00000172747.8
ENSMUST00000105801.9 ENSMUST00000053862.12 |
Slc66a1
|
solute carrier family 66 member 1 |
| chr7_+_79675727 | 0.42 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr18_+_60426444 | 0.41 |
ENSMUST00000171297.2
|
F830016B08Rik
|
RIKEN cDNA F830016B08 gene |
| chr8_-_116434517 | 0.40 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr13_+_55097200 | 0.40 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr1_+_87254729 | 0.37 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr8_+_3410842 | 0.37 |
ENSMUST00000238813.2
|
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18 |
| chr16_+_43330630 | 0.36 |
ENSMUST00000114695.3
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_+_87185159 | 0.36 |
ENSMUST00000215163.3
|
Olfr1120
|
olfactory receptor 1120 |
| chr15_-_102630589 | 0.36 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr6_-_70237939 | 0.35 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr11_+_77239098 | 0.35 |
ENSMUST00000181283.3
|
Ssh2
|
slingshot protein phosphatase 2 |
| chr6_-_57372768 | 0.34 |
ENSMUST00000228574.2
|
Vmn1r18
|
vomeronasal 1 receptor 18 |
| chr2_+_48839505 | 0.33 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr4_-_156312961 | 0.33 |
ENSMUST00000217885.2
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr6_-_58195806 | 0.32 |
ENSMUST00000228530.2
ENSMUST00000226666.2 |
Vmn1r27
|
vomeronasal 1 receptor 27 |
| chr14_-_45456821 | 0.31 |
ENSMUST00000128484.8
ENSMUST00000147853.8 ENSMUST00000022377.11 ENSMUST00000143609.2 ENSMUST00000153383.8 ENSMUST00000139526.9 |
Txndc16
|
thioredoxin domain containing 16 |
| chr19_-_4447080 | 0.31 |
ENSMUST00000075856.11
ENSMUST00000176483.3 ENSMUST00000116571.9 |
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr10_-_80578057 | 0.30 |
ENSMUST00000020420.9
|
Ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr2_-_104542467 | 0.30 |
ENSMUST00000111118.8
ENSMUST00000028597.4 |
Tcp11l1
|
t-complex 11 like 1 |
| chr7_-_104991477 | 0.28 |
ENSMUST00000213290.2
|
Olfr691
|
olfactory receptor 691 |
| chr17_+_27171834 | 0.28 |
ENSMUST00000231853.2
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr2_+_97298002 | 0.28 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr6_-_36787096 | 0.28 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr15_+_25622611 | 0.27 |
ENSMUST00000110457.8
ENSMUST00000137601.8 |
Myo10
|
myosin X |
| chr6_+_58278534 | 0.27 |
ENSMUST00000228909.2
ENSMUST00000227761.2 ENSMUST00000228038.2 ENSMUST00000226971.2 |
Vmn1r29
|
vomeronasal 1 receptor 29 |
| chrX_+_167819606 | 0.26 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chrX_+_100492684 | 0.26 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr17_+_75772475 | 0.25 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr4_-_156312996 | 0.25 |
ENSMUST00000105571.4
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr2_-_73410632 | 0.24 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr4_-_43040278 | 0.24 |
ENSMUST00000107958.8
ENSMUST00000107959.8 ENSMUST00000152846.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chr15_+_61858883 | 0.24 |
ENSMUST00000159338.2
|
Myc
|
myelocytomatosis oncogene |
| chr11_+_82842921 | 0.23 |
ENSMUST00000108158.9
ENSMUST00000067443.10 |
Slfn5
|
schlafen 5 |
| chr11_+_110858842 | 0.23 |
ENSMUST00000180023.8
ENSMUST00000106636.8 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr2_+_37406630 | 0.23 |
ENSMUST00000065441.7
|
Gpr21
|
G protein-coupled receptor 21 |
| chr10_+_127337541 | 0.22 |
ENSMUST00000160019.8
ENSMUST00000160610.2 ENSMUST00000035839.3 |
Stac3
|
SH3 and cysteine rich domain 3 |
| chr14_+_45457168 | 0.22 |
ENSMUST00000227086.2
ENSMUST00000147957.2 |
Gpr137c
|
G protein-coupled receptor 137C |
| chr3_-_88363704 | 0.22 |
ENSMUST00000141471.2
ENSMUST00000123753.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr11_+_85243970 | 0.22 |
ENSMUST00000108056.8
ENSMUST00000108061.8 ENSMUST00000108062.8 ENSMUST00000138423.8 ENSMUST00000092821.10 ENSMUST00000074875.11 |
Bcas3
|
breast carcinoma amplified sequence 3 |
| chr9_+_56979307 | 0.21 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr6_-_66620810 | 0.21 |
ENSMUST00000228647.2
ENSMUST00000227332.2 ENSMUST00000226262.2 ENSMUST00000226999.2 |
Vmn1r34
|
vomeronasal 1 receptor 34 |
| chr9_+_108174052 | 0.20 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr1_+_171052623 | 0.20 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr8_-_37200051 | 0.20 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr9_-_51076724 | 0.19 |
ENSMUST00000210433.2
|
Gm32742
|
predicted gene, 32742 |
| chr19_+_45036037 | 0.19 |
ENSMUST00000062213.13
|
Sfxn3
|
sideroflexin 3 |
| chr5_-_90031180 | 0.19 |
ENSMUST00000198151.2
|
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3 |
| chr2_-_148574353 | 0.19 |
ENSMUST00000028926.13
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chr8_-_84299540 | 0.19 |
ENSMUST00000212990.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr14_+_20757615 | 0.18 |
ENSMUST00000022358.9
ENSMUST00000224751.2 |
Zswim8
|
zinc finger SWIM-type containing 8 |
| chr6_-_124519240 | 0.18 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr16_+_17269845 | 0.18 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr18_-_89787603 | 0.18 |
ENSMUST00000097495.5
|
Dok6
|
docking protein 6 |
| chr4_+_122910382 | 0.17 |
ENSMUST00000102649.4
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr1_-_121255448 | 0.17 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr7_+_4925781 | 0.17 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr1_-_79739469 | 0.17 |
ENSMUST00000113512.8
ENSMUST00000113513.8 ENSMUST00000113515.8 ENSMUST00000113514.8 ENSMUST00000113510.8 ENSMUST00000113511.8 ENSMUST00000048820.14 |
Wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr19_-_5560473 | 0.16 |
ENSMUST00000237111.2
ENSMUST00000070172.6 |
Snx32
|
sorting nexin 32 |
| chr11_-_115977755 | 0.16 |
ENSMUST00000074628.13
ENSMUST00000106444.4 |
Wbp2
|
WW domain binding protein 2 |
| chr10_+_60182630 | 0.16 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr8_+_3550450 | 0.16 |
ENSMUST00000004683.13
ENSMUST00000160338.2 |
Mcoln1
|
mucolipin 1 |
| chr19_+_34585322 | 0.16 |
ENSMUST00000076249.6
|
Ifit3b
|
interferon-induced protein with tetratricopeptide repeats 3B |
| chr9_+_64142483 | 0.16 |
ENSMUST00000039011.4
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr9_-_21763898 | 0.15 |
ENSMUST00000217336.2
ENSMUST00000034728.9 |
Dock6
|
dedicator of cytokinesis 6 |
| chr15_-_82796308 | 0.15 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr2_-_63928339 | 0.15 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr2_-_118380149 | 0.15 |
ENSMUST00000090219.13
|
Bmf
|
BCL2 modifying factor |
| chr7_-_30259025 | 0.15 |
ENSMUST00000043975.11
ENSMUST00000156241.2 |
Lin37
|
lin-37 homolog (C. elegans) |
| chr5_-_131567526 | 0.14 |
ENSMUST00000161374.8
|
Auts2
|
autism susceptibility candidate 2 |
| chr19_+_45035942 | 0.14 |
ENSMUST00000237222.2
ENSMUST00000111954.11 |
Sfxn3
|
sideroflexin 3 |
| chr11_-_114851243 | 0.14 |
ENSMUST00000092466.13
ENSMUST00000061637.4 |
Cd300c
|
CD300C molecule |
| chr18_-_10610332 | 0.14 |
ENSMUST00000025142.13
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr2_-_77000936 | 0.14 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr6_+_92846338 | 0.14 |
ENSMUST00000113434.2
|
Gm15737
|
predicted gene 15737 |
| chr11_-_75345482 | 0.14 |
ENSMUST00000173320.8
|
Wdr81
|
WD repeat domain 81 |
| chr8_+_66838927 | 0.14 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr6_-_70412460 | 0.13 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chr14_+_30601157 | 0.13 |
ENSMUST00000040715.8
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr18_+_4994600 | 0.13 |
ENSMUST00000140448.8
|
Svil
|
supervillin |
| chr1_-_80642969 | 0.13 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chr18_+_42408418 | 0.13 |
ENSMUST00000091920.6
ENSMUST00000046972.14 ENSMUST00000236240.2 |
Rbm27
|
RNA binding motif protein 27 |
| chr6_-_142647944 | 0.13 |
ENSMUST00000100827.5
ENSMUST00000087527.11 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr3_-_79439181 | 0.12 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2 |
| chr4_-_135749032 | 0.12 |
ENSMUST00000030427.6
|
Eloa
|
elongin A |
| chr14_+_78141679 | 0.12 |
ENSMUST00000022591.16
ENSMUST00000169978.2 ENSMUST00000227903.2 |
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr11_+_68582923 | 0.12 |
ENSMUST00000018887.15
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr7_-_30251680 | 0.11 |
ENSMUST00000215288.2
ENSMUST00000108165.8 ENSMUST00000153594.3 |
Proser3
|
proline and serine rich 3 |
| chr6_-_58452315 | 0.11 |
ENSMUST00000177318.3
ENSMUST00000176147.9 ENSMUST00000176023.3 ENSMUST00000228586.2 |
Vmn1r31
|
vomeronasal 1 receptor 31 |
| chr13_+_64309675 | 0.11 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr7_+_101032021 | 0.11 |
ENSMUST00000141083.9
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr4_-_134431534 | 0.11 |
ENSMUST00000054096.13
ENSMUST00000038628.4 |
Man1c1
|
mannosidase, alpha, class 1C, member 1 |
| chr9_-_37166699 | 0.11 |
ENSMUST00000161114.2
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chrX_+_47197789 | 0.11 |
ENSMUST00000114998.8
ENSMUST00000115000.4 |
Xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr2_-_45007407 | 0.11 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr15_-_71599664 | 0.11 |
ENSMUST00000022953.10
|
Fam135b
|
family with sequence similarity 135, member B |
| chr2_+_30485048 | 0.11 |
ENSMUST00000102853.4
|
Cstad
|
CSA-conditional, T cell activation-dependent protein |
| chr11_-_60804601 | 0.10 |
ENSMUST00000019075.4
|
Natd1
|
N-acetyltransferase domain containing 1 |
| chr2_+_32253692 | 0.10 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr1_-_121255400 | 0.10 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr1_+_87133521 | 0.10 |
ENSMUST00000188796.7
ENSMUST00000027470.14 ENSMUST00000186038.2 |
Chrng
|
cholinergic receptor, nicotinic, gamma polypeptide |
| chr14_-_56809287 | 0.10 |
ENSMUST00000065302.8
|
Cenpj
|
centromere protein J |
| chr4_+_136150835 | 0.10 |
ENSMUST00000088677.5
ENSMUST00000121571.8 ENSMUST00000117699.2 |
Htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D |
| chr19_-_8816530 | 0.10 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr6_+_126916919 | 0.10 |
ENSMUST00000032497.7
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr17_+_33774681 | 0.10 |
ENSMUST00000087605.13
ENSMUST00000174695.2 |
Myo1f
|
myosin IF |
| chr3_-_84128160 | 0.10 |
ENSMUST00000154152.2
ENSMUST00000107693.9 ENSMUST00000107695.9 |
Trim2
|
tripartite motif-containing 2 |
| chr6_-_142647985 | 0.10 |
ENSMUST00000205202.3
ENSMUST00000073173.12 ENSMUST00000111771.8 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr1_+_128069677 | 0.10 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr10_+_75729237 | 0.09 |
ENSMUST00000009236.6
ENSMUST00000217811.2 |
Derl3
|
Der1-like domain family, member 3 |
| chr6_-_97156032 | 0.09 |
ENSMUST00000095664.6
|
Tmf1
|
TATA element modulatory factor 1 |
| chr7_-_141015240 | 0.09 |
ENSMUST00000138865.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr5_+_73071897 | 0.09 |
ENSMUST00000200785.2
|
Slain2
|
SLAIN motif family, member 2 |
| chr9_-_45896110 | 0.09 |
ENSMUST00000215060.2
ENSMUST00000213853.2 ENSMUST00000216334.2 |
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr1_-_80687213 | 0.09 |
ENSMUST00000186087.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr3_-_95214102 | 0.09 |
ENSMUST00000107183.8
ENSMUST00000164406.8 ENSMUST00000123365.2 |
Anxa9
|
annexin A9 |
| chr10_-_117546660 | 0.09 |
ENSMUST00000020408.16
ENSMUST00000105263.9 |
Mdm2
|
transformed mouse 3T3 cell double minute 2 |
| chr3_-_95214443 | 0.09 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr14_+_67470884 | 0.09 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr10_+_60113449 | 0.09 |
ENSMUST00000105465.8
ENSMUST00000179238.8 ENSMUST00000177779.8 ENSMUST00000004316.15 |
Psap
|
prosaposin |
| chr7_-_23907518 | 0.08 |
ENSMUST00000086006.12
|
Zfp111
|
zinc finger protein 111 |
| chr11_-_100713348 | 0.08 |
ENSMUST00000107358.9
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr6_-_53797748 | 0.08 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr13_+_44884200 | 0.08 |
ENSMUST00000173704.8
ENSMUST00000044608.14 ENSMUST00000173367.8 |
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr17_+_30223707 | 0.08 |
ENSMUST00000226208.3
|
Zfand3
|
zinc finger, AN1-type domain 3 |
| chr15_-_75620060 | 0.08 |
ENSMUST00000062002.6
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr15_-_102630496 | 0.08 |
ENSMUST00000171838.2
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr5_-_137246852 | 0.08 |
ENSMUST00000179412.2
|
Muc3a
|
mucin 3A, cell surface associated |
| chr5_-_137246611 | 0.08 |
ENSMUST00000196391.5
|
Muc3a
|
mucin 3A, cell surface associated |
| chr15_-_99425555 | 0.08 |
ENSMUST00000231171.2
|
Faim2
|
Fas apoptotic inhibitory molecule 2 |
| chr8_+_3550533 | 0.08 |
ENSMUST00000208306.2
|
Mcoln1
|
mucolipin 1 |
| chr6_-_70121150 | 0.08 |
ENSMUST00000197525.2
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr19_-_40502117 | 0.08 |
ENSMUST00000225148.2
ENSMUST00000224667.2 ENSMUST00000225153.2 ENSMUST00000238831.2 ENSMUST00000226047.2 ENSMUST00000224247.2 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr18_+_38430015 | 0.08 |
ENSMUST00000236319.2
|
Rnf14
|
ring finger protein 14 |
| chr5_-_34671236 | 0.07 |
ENSMUST00000114359.2
ENSMUST00000030991.14 ENSMUST00000087737.10 |
Tnip2
|
TNFAIP3 interacting protein 2 |
| chr10_+_128106414 | 0.07 |
ENSMUST00000085708.3
ENSMUST00000105238.10 |
Stat2
|
signal transducer and activator of transcription 2 |
| chr1_-_9818601 | 0.07 |
ENSMUST00000057438.7
|
Vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr9_+_74883377 | 0.07 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr5_+_134128543 | 0.07 |
ENSMUST00000016088.9
|
Castor2
|
cytosolic arginine sensor for mTORC1 subunit 2 |
| chr14_+_54496683 | 0.07 |
ENSMUST00000041197.13
ENSMUST00000239403.2 ENSMUST00000197605.3 |
Abhd4
|
abhydrolase domain containing 4 |
| chr17_+_29768757 | 0.07 |
ENSMUST00000048677.9
ENSMUST00000150388.3 |
Tbc1d22b
Gm28052
|
TBC1 domain family, member 22B predicted gene, 28052 |
| chr4_+_74170165 | 0.07 |
ENSMUST00000030102.12
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr9_+_75093177 | 0.07 |
ENSMUST00000129281.8
ENSMUST00000148144.8 ENSMUST00000130384.2 |
Myo5a
|
myosin VA |
| chr4_+_128777339 | 0.07 |
ENSMUST00000035667.9
|
Trim62
|
tripartite motif-containing 62 |
| chr1_-_153726930 | 0.07 |
ENSMUST00000059607.8
|
Teddm2
|
transmembrane epididymal family member 2 |
| chr15_+_61857226 | 0.07 |
ENSMUST00000161976.8
ENSMUST00000022971.8 |
Myc
|
myelocytomatosis oncogene |
| chr6_+_136530970 | 0.07 |
ENSMUST00000189535.2
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr5_+_14564932 | 0.07 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr14_-_36628263 | 0.06 |
ENSMUST00000183007.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_-_36597313 | 0.06 |
ENSMUST00000195339.3
|
Sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr5_-_18093739 | 0.06 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr2_+_170573727 | 0.06 |
ENSMUST00000029075.5
|
Dok5
|
docking protein 5 |
| chr13_+_44884757 | 0.06 |
ENSMUST00000174086.2
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr8_+_84699580 | 0.06 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr10_+_24025188 | 0.06 |
ENSMUST00000020174.7
ENSMUST00000220041.2 |
Stx7
|
syntaxin 7 |
| chr8_+_22550727 | 0.06 |
ENSMUST00000072572.13
ENSMUST00000110737.3 ENSMUST00000131624.2 |
Alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr6_-_54543446 | 0.06 |
ENSMUST00000019268.11
|
Scrn1
|
secernin 1 |
| chr6_-_70435020 | 0.06 |
ENSMUST00000198184.2
|
Igkv6-13
|
immunoglobulin kappa variable 6-13 |
| chr2_-_143704853 | 0.06 |
ENSMUST00000099296.4
|
Bfsp1
|
beaded filament structural protein 1, in lens-CP94 |
| chr6_-_70094604 | 0.06 |
ENSMUST00000103378.3
|
Igkv8-30
|
immunoglobulin kappa chain variable 8-30 |
| chr2_-_150327011 | 0.06 |
ENSMUST00000109914.2
|
Zfp345
|
zinc finger protein 345 |
| chr9_-_45895635 | 0.06 |
ENSMUST00000215427.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr5_-_147662798 | 0.06 |
ENSMUST00000110529.6
ENSMUST00000031653.12 |
Flt1
|
FMS-like tyrosine kinase 1 |
| chr1_-_80643024 | 0.05 |
ENSMUST00000187774.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr1_-_134260666 | 0.05 |
ENSMUST00000168515.8
ENSMUST00000189361.2 |
Ppfia4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr9_-_63509699 | 0.05 |
ENSMUST00000171243.2
ENSMUST00000163982.8 ENSMUST00000163624.8 |
Iqch
|
IQ motif containing H |
| chr17_+_30224003 | 0.05 |
ENSMUST00000057897.10
|
Zfand3
|
zinc finger, AN1-type domain 3 |
| chr1_+_88154727 | 0.05 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr8_-_36716445 | 0.05 |
ENSMUST00000239119.2
ENSMUST00000065297.6 |
Lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr5_+_108536242 | 0.05 |
ENSMUST00000031456.8
|
Pde6b
|
phosphodiesterase 6B, cGMP, rod receptor, beta polypeptide |
| chr11_+_97253221 | 0.05 |
ENSMUST00000238729.2
ENSMUST00000045540.4 |
Socs7
|
suppressor of cytokine signaling 7 |
| chr3_+_95341698 | 0.05 |
ENSMUST00000102749.11
ENSMUST00000090804.12 ENSMUST00000107161.8 ENSMUST00000107160.8 ENSMUST00000015666.17 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr14_+_22069877 | 0.04 |
ENSMUST00000161249.8
ENSMUST00000159777.8 ENSMUST00000162540.2 |
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr11_-_68762664 | 0.04 |
ENSMUST00000101017.9
|
Ndel1
|
nudE neurodevelopment protein 1 like 1 |
| chr8_-_47742389 | 0.04 |
ENSMUST00000211737.2
|
Stox2
|
storkhead box 2 |
| chr11_-_120534469 | 0.04 |
ENSMUST00000141254.8
ENSMUST00000170556.8 ENSMUST00000151876.8 ENSMUST00000026133.15 ENSMUST00000139706.2 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr5_-_146521629 | 0.04 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr2_-_169973076 | 0.04 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr9_-_63509747 | 0.04 |
ENSMUST00000080527.12
ENSMUST00000042322.11 |
Iqch
|
IQ motif containing H |
| chr13_+_44884740 | 0.03 |
ENSMUST00000173246.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr3_-_108352499 | 0.03 |
ENSMUST00000090553.12
ENSMUST00000153499.2 |
Sars
|
seryl-aminoacyl-tRNA synthetase |
| chr7_+_30411634 | 0.03 |
ENSMUST00000005692.14
ENSMUST00000170371.2 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr6_-_70120881 | 0.03 |
ENSMUST00000103380.3
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr3_-_108062172 | 0.03 |
ENSMUST00000062028.8
|
Gpr61
|
G protein-coupled receptor 61 |
| chr2_-_160208977 | 0.03 |
ENSMUST00000099126.5
|
Mafb
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
| chr1_-_190950129 | 0.03 |
ENSMUST00000195117.2
|
Atf3
|
activating transcription factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.2 | 0.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.6 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 0.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.3 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.3 | GO:0090096 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.3 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0046340 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.0 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.1 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.0 | 0.7 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.2 | GO:0034145 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 0.0 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.0 | 0.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.6 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.5 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |